Probe CUST_52214_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52214_PI426222305 | JHI_St_60k_v1 | DMT400075154 | CAGCCAATTGTTTCTGGTAATAGTTTTCCCCTTTTGAGTGTCTGTCTGTTCTTCCTTGTT |
All Microarray Probes Designed to Gene DMG400029237
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52215_PI426222305 | JHI_St_60k_v1 | DMT400075156 | ATGAAGGCTGAATCGGAACCGGAAAAGGCACAGATGACGATATTCTATGCAGATTTACCA |
| CUST_52218_PI426222305 | JHI_St_60k_v1 | DMT400075153 | GGTTGTTCAATTCATCATCTCTCAAGTTTTCATGTGTTCGCAACAACAATTTATCTGTCT |
| CUST_52217_PI426222305 | JHI_St_60k_v1 | DMT400075155 | GGTTGTTCAATTCATCATCTCTCAAGTTTTCATGTGTTCGCAACAACAATTTATCTGTCT |
| CUST_52214_PI426222305 | JHI_St_60k_v1 | DMT400075154 | CAGCCAATTGTTTCTGGTAATAGTTTTCCCCTTTTGAGTGTCTGTCTGTTCTTCCTTGTT |
Microarray Signals from CUST_52214_PI426222305
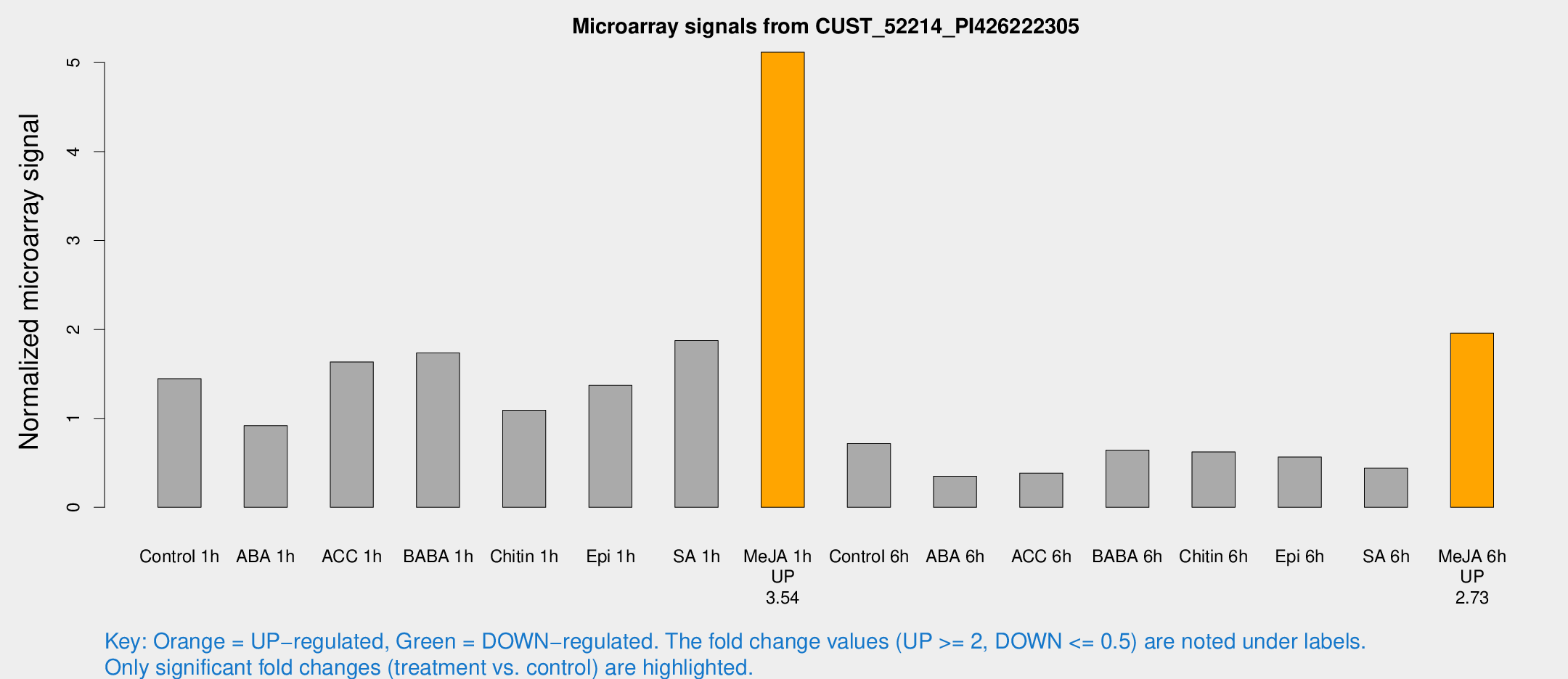
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 766.2 | 156.162 | 1.44557 | 0.207852 |
| ABA 1h | 445.937 | 116.562 | 0.917456 | 0.19822 |
| ACC 1h | 970.976 | 288.771 | 1.63488 | 0.536001 |
| BABA 1h | 884.417 | 159.173 | 1.73617 | 0.169623 |
| Chitin 1h | 501.408 | 29.172 | 1.09111 | 0.119637 |
| Epi 1h | 607.948 | 39.9164 | 1.37058 | 0.0804814 |
| SA 1h | 996.092 | 110.394 | 1.87534 | 0.339207 |
| Me-JA 1h | 2172.28 | 306.583 | 5.11693 | 0.512028 |
| Control 6h | 401.387 | 112.501 | 0.717205 | 0.161766 |
| ABA 6h | 190.993 | 15.2839 | 0.349897 | 0.0220885 |
| ACC 6h | 235.326 | 47.2059 | 0.384354 | 0.066549 |
| BABA 6h | 374.285 | 48.4408 | 0.643883 | 0.0610697 |
| Chitin 6h | 339.599 | 20.0356 | 0.623375 | 0.0396357 |
| Epi 6h | 350.294 | 94.7587 | 0.564726 | 0.191392 |
| SA 6h | 238.944 | 66.9786 | 0.440139 | 0.0896704 |
| Me-JA 6h | 1028.4 | 173.238 | 1.95844 | 0.235251 |
Source Transcript PGSC0003DMT400075154 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G74950.1 | +3 | 2e-22 | 76 | 65/186 (35%) | TIFY domain/Divergent CCT motif family protein | chr1:28148919-28150258 REVERSE LENGTH=249 |
