Probe CUST_51712_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51712_PI426222305 | JHI_St_60k_v1 | DMT400028480 | CAGAACTCAGTGTCTCACAATGAAATGCATTATGACAGAAAGGGAAGTTTGTAACTTTTA |
All Microarray Probes Designed to Gene DMG400010963
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51710_PI426222305 | JHI_St_60k_v1 | DMT400028481 | CAGAACTCAGTGTCTCACAATGAAATGCATTATGACAGAAAGGGAAGTTTGTAACTTTTA |
| CUST_51709_PI426222305 | JHI_St_60k_v1 | DMT400028482 | CAGAACTCAGTGTCTCACAATGAAATGCATTATGACAGAAAGGGAAGTTTGTAACTTTTA |
| CUST_51712_PI426222305 | JHI_St_60k_v1 | DMT400028480 | CAGAACTCAGTGTCTCACAATGAAATGCATTATGACAGAAAGGGAAGTTTGTAACTTTTA |
Microarray Signals from CUST_51712_PI426222305
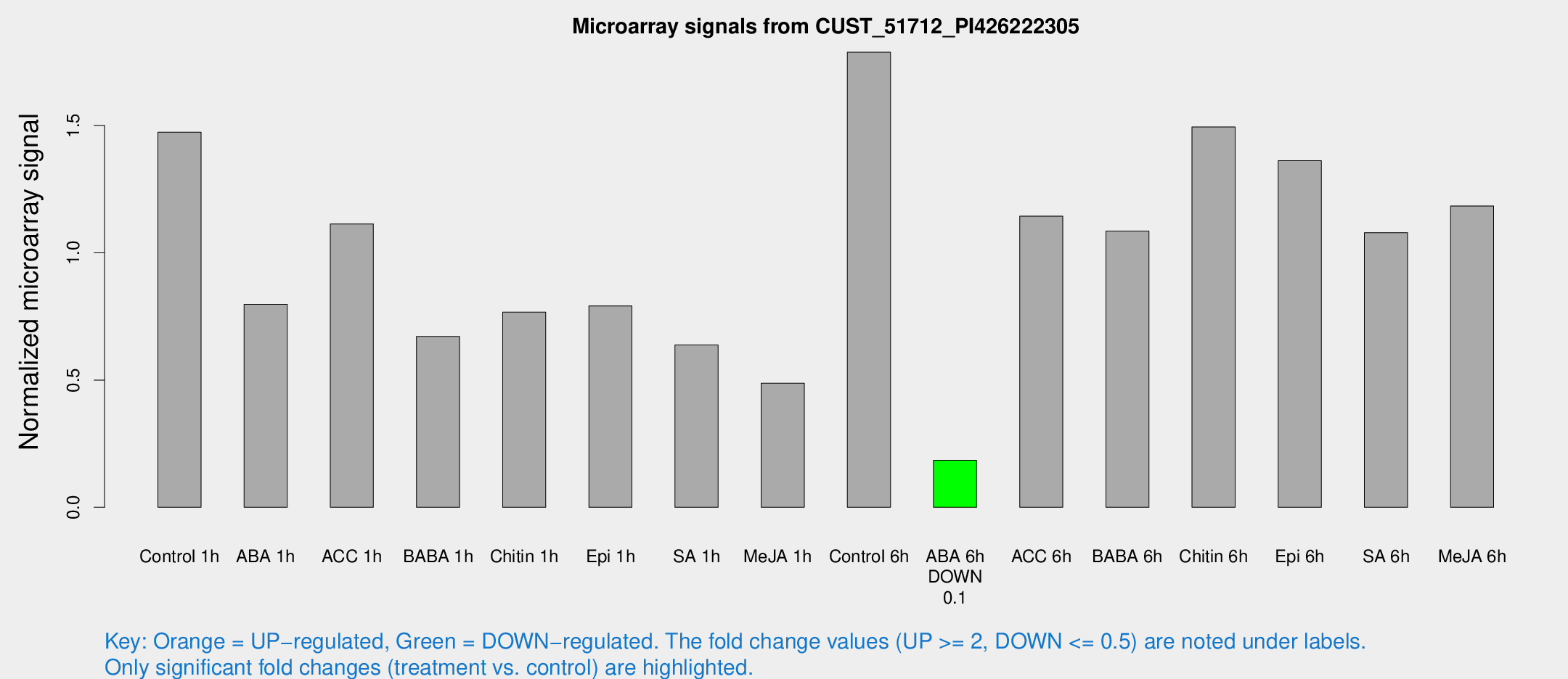
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 506.913 | 91.4656 | 1.47375 | 0.19564 |
| ABA 1h | 237.648 | 24.231 | 0.797307 | 0.125317 |
| ACC 1h | 388.711 | 51.1971 | 1.11304 | 0.0691381 |
| BABA 1h | 221.958 | 34.8098 | 0.671031 | 0.0452121 |
| Chitin 1h | 240.134 | 50.7747 | 0.766864 | 0.152334 |
| Epi 1h | 231.296 | 25.8624 | 0.791313 | 0.0676505 |
| SA 1h | 220.481 | 21.0141 | 0.637788 | 0.0788133 |
| Me-JA 1h | 136.455 | 23.4371 | 0.487579 | 0.0899357 |
| Control 6h | 651.271 | 165.867 | 1.78781 | 0.40414 |
| ABA 6h | 66.3545 | 7.78492 | 0.184543 | 0.0178183 |
| ACC 6h | 451.393 | 79.4021 | 1.1438 | 0.0695525 |
| BABA 6h | 418.525 | 72.9737 | 1.0851 | 0.227755 |
| Chitin 6h | 533.557 | 38.7462 | 1.49422 | 0.113137 |
| Epi 6h | 538.288 | 118.687 | 1.36198 | 0.377076 |
| SA 6h | 372.873 | 73.466 | 1.07878 | 0.116247 |
| Me-JA 6h | 406.69 | 69.507 | 1.18398 | 0.128052 |
Source Transcript PGSC0003DMT400028480 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G79670.1 | +1 | 3e-70 | 237 | 125/263 (48%) | Wall-associated kinase family protein | chr1:29976887-29979337 REVERSE LENGTH=751 |
