Probe CUST_51136_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51136_PI426222305 | JHI_St_60k_v1 | DMT400019823 | GAGGACACATAGATGGCTAAATGCAAGGCATGTAATCATGTTTATGTACAAAAGTCAATA |
All Microarray Probes Designed to Gene DMG400007665
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51132_PI426222305 | JHI_St_60k_v1 | DMT400019824 | CTCCACCTCAAAGGTATACTAGTACACCTCATTTTTATTCATTTGGTTTATGGATAGAGT |
| CUST_51136_PI426222305 | JHI_St_60k_v1 | DMT400019823 | GAGGACACATAGATGGCTAAATGCAAGGCATGTAATCATGTTTATGTACAAAAGTCAATA |
Microarray Signals from CUST_51136_PI426222305
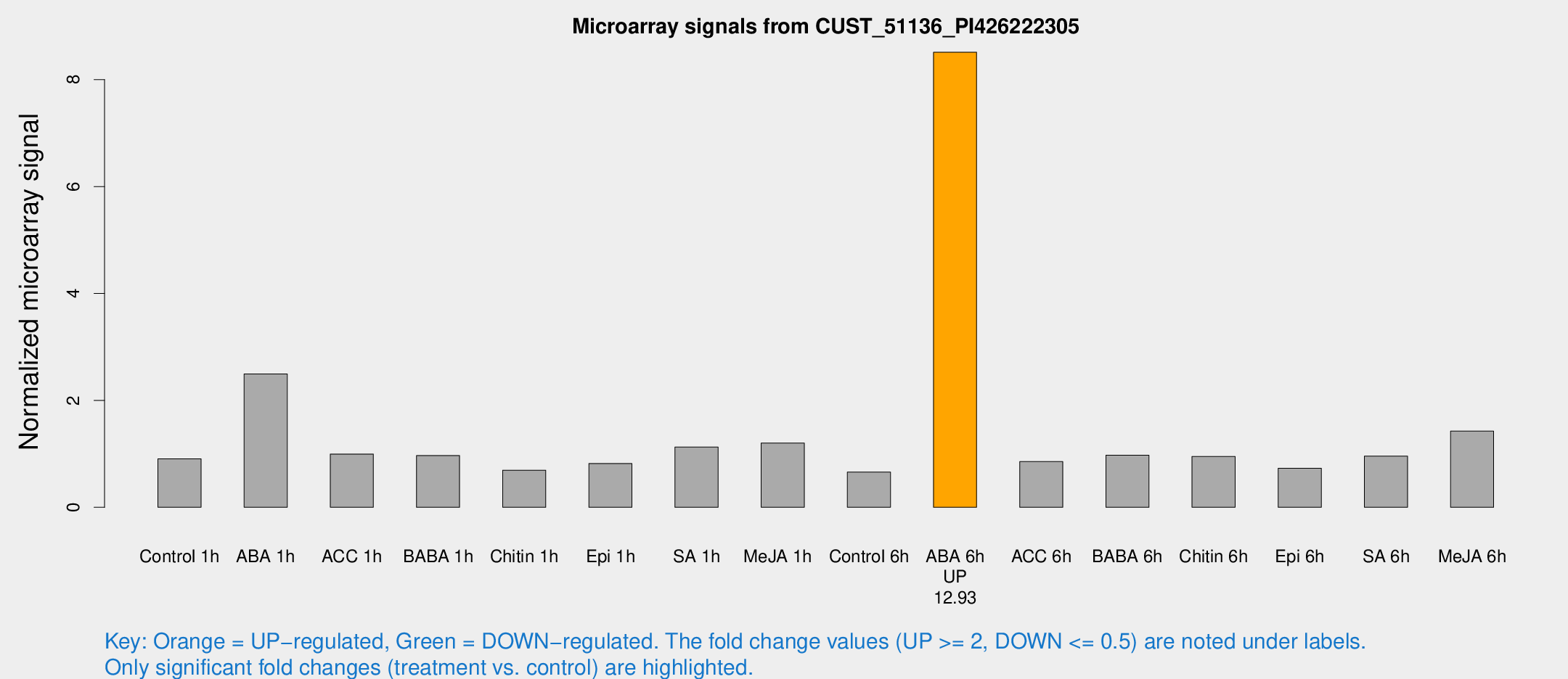
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1573.17 | 170.584 | 0.906124 | 0.137721 |
| ABA 1h | 3944.88 | 787.756 | 2.49362 | 0.463846 |
| ACC 1h | 2055.28 | 697.887 | 0.996681 | 0.3883 |
| BABA 1h | 1622.51 | 179.014 | 0.968661 | 0.0559681 |
| Chitin 1h | 1086.17 | 123.182 | 0.694463 | 0.0936404 |
| Epi 1h | 1285.65 | 277.914 | 0.819541 | 0.199563 |
| SA 1h | 1990.55 | 115.292 | 1.12648 | 0.0811758 |
| Me-JA 1h | 1697.75 | 170.382 | 1.20079 | 0.0693695 |
| Control 6h | 1244.08 | 331.004 | 0.658449 | 0.158987 |
| ABA 6h | 16677.2 | 3971.94 | 8.5125 | 2.07739 |
| ACC 6h | 1693.87 | 134.248 | 0.855571 | 0.0672842 |
| BABA 6h | 1932.17 | 355.314 | 0.975476 | 0.145521 |
| Chitin 6h | 1741.23 | 113.326 | 0.949422 | 0.0915417 |
| Epi 6h | 1431.13 | 166.356 | 0.730186 | 0.0732527 |
| SA 6h | 1626.46 | 94.0993 | 0.95736 | 0.124363 |
| Me-JA 6h | 2474.36 | 319.425 | 1.42491 | 0.0822929 |
Source Transcript PGSC0003DMT400019823 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G23910.1 | +3 | 3e-78 | 248 | 138/290 (48%) | NAD(P)-binding Rossmann-fold superfamily protein | chr2:10177902-10179789 FORWARD LENGTH=304 |
