Probe CUST_51132_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51132_PI426222305 | JHI_St_60k_v1 | DMT400019824 | CTCCACCTCAAAGGTATACTAGTACACCTCATTTTTATTCATTTGGTTTATGGATAGAGT |
All Microarray Probes Designed to Gene DMG400007665
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51132_PI426222305 | JHI_St_60k_v1 | DMT400019824 | CTCCACCTCAAAGGTATACTAGTACACCTCATTTTTATTCATTTGGTTTATGGATAGAGT |
| CUST_51136_PI426222305 | JHI_St_60k_v1 | DMT400019823 | GAGGACACATAGATGGCTAAATGCAAGGCATGTAATCATGTTTATGTACAAAAGTCAATA |
Microarray Signals from CUST_51132_PI426222305
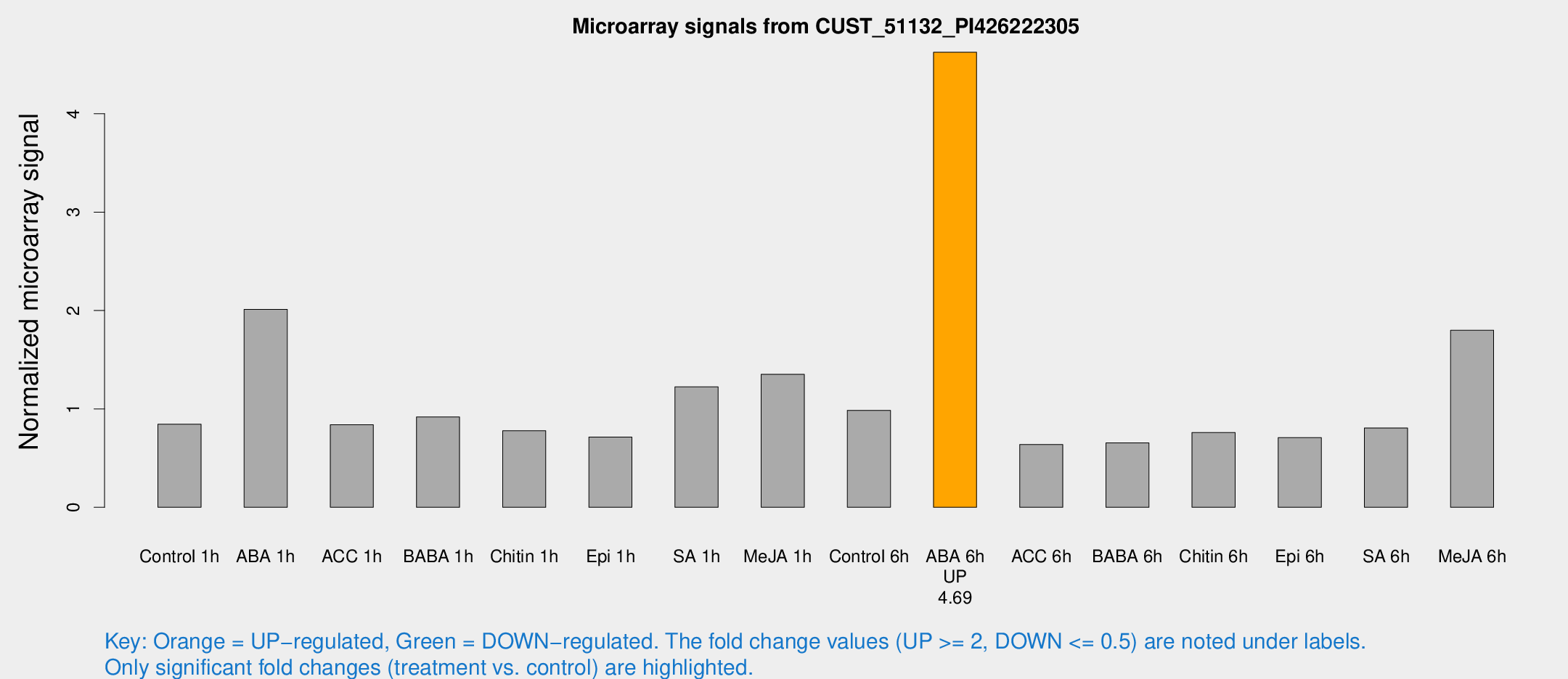
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 36.304 | 4.00041 | 0.844248 | 0.13041 |
| ABA 1h | 81.8001 | 20.4101 | 2.01152 | 0.526174 |
| ACC 1h | 43.5192 | 14.8193 | 0.838473 | 0.357574 |
| BABA 1h | 39.1875 | 7.29329 | 0.91853 | 0.103221 |
| Chitin 1h | 30.8076 | 6.10127 | 0.778078 | 0.0943214 |
| Epi 1h | 27.2827 | 5.53403 | 0.714437 | 0.151819 |
| SA 1h | 54.9433 | 8.65429 | 1.2239 | 0.154424 |
| Me-JA 1h | 47.9912 | 7.68566 | 1.35122 | 0.142561 |
| Control 6h | 42.9546 | 6.20743 | 0.985824 | 0.0984344 |
| ABA 6h | 226.503 | 57.6609 | 4.62584 | 1.28343 |
| ACC 6h | 33.0454 | 7.84331 | 0.637547 | 0.198884 |
| BABA 6h | 34.9779 | 11.7953 | 0.654317 | 0.214947 |
| Chitin 6h | 34.3543 | 4.09999 | 0.759458 | 0.0906954 |
| Epi 6h | 35.0577 | 6.67826 | 0.708019 | 0.128786 |
| SA 6h | 34.4826 | 4.5224 | 0.805682 | 0.170702 |
| Me-JA 6h | 76.2313 | 5.46609 | 1.79985 | 0.157018 |
Source Transcript PGSC0003DMT400019824 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G23910.1 | +3 | 4e-79 | 247 | 131/266 (49%) | NAD(P)-binding Rossmann-fold superfamily protein | chr2:10177902-10179789 FORWARD LENGTH=304 |
