Probe CUST_5053_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5053_PI426222305 | JHI_St_60k_v1 | DMT400009313 | GTCTGACAAGAAAGTGGTTCTTTTAACAGCTAAACAATATCGAAGAAAGCATTCAGTTCA |
All Microarray Probes Designed to Gene DMG400003619
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5182_PI426222305 | JHI_St_60k_v1 | DMT400009311 | TGGGCTTATACTGCTCTTGCTGGGGTTGAGAGGAATATTCAGAACAAACAGAAGGCAGTA |
| CUST_4978_PI426222305 | JHI_St_60k_v1 | DMT400009314 | GTCACTGCTCTCTCCAGTGAGAGTGCGAAACTAATTAAGAATTATTATTTATATCCAGAA |
| CUST_5053_PI426222305 | JHI_St_60k_v1 | DMT400009313 | GTCTGACAAGAAAGTGGTTCTTTTAACAGCTAAACAATATCGAAGAAAGCATTCAGTTCA |
| CUST_5121_PI426222305 | JHI_St_60k_v1 | DMT400009312 | GTCACTGCTCTCTCCAGTGAGAGTGCGAAACTAATTAAGAATTATTATTTATATCCAGAA |
Microarray Signals from CUST_5053_PI426222305
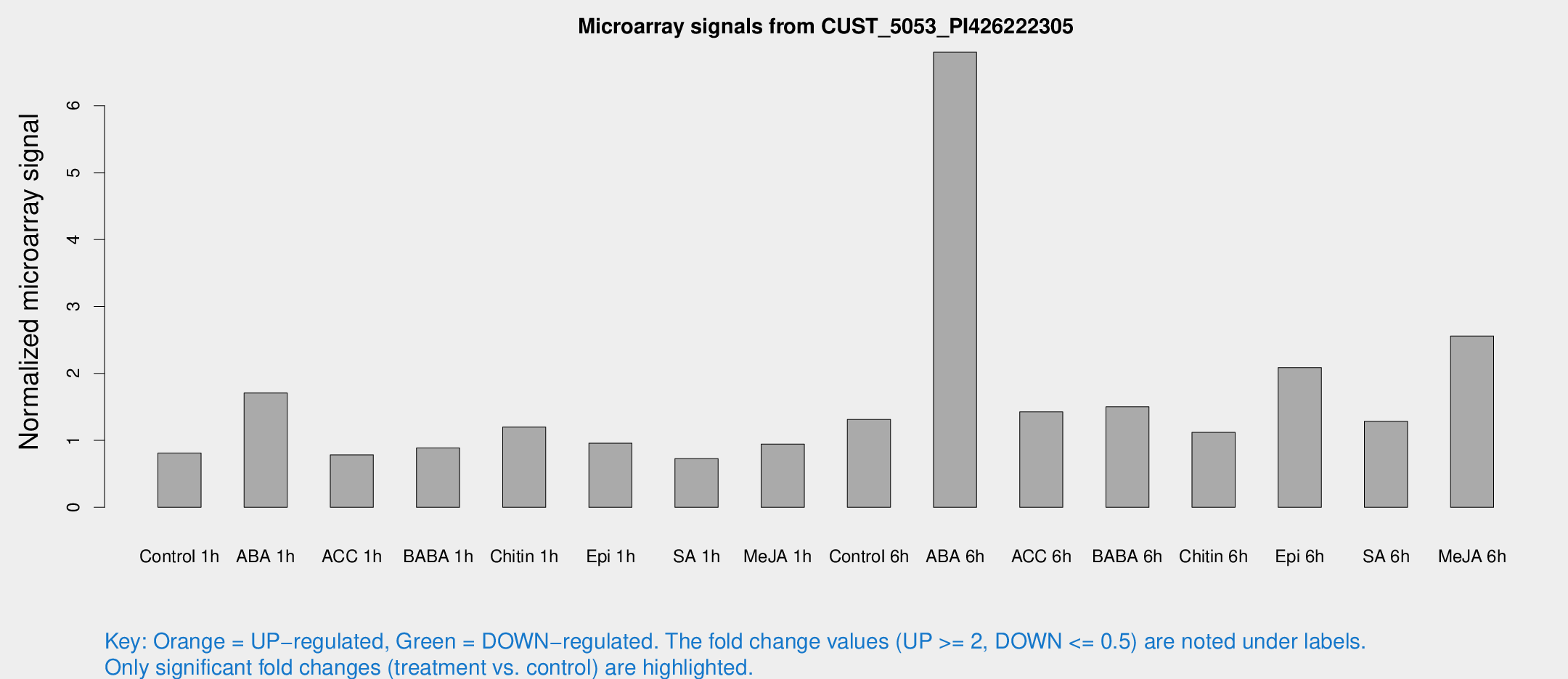
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.15253 | 3.27604 | 0.811407 | 0.438762 |
| ABA 1h | 12.6509 | 3.544 | 1.70798 | 0.818907 |
| ACC 1h | 6.04422 | 3.50247 | 0.78332 | 0.453644 |
| BABA 1h | 6.44949 | 3.49248 | 0.887455 | 0.484179 |
| Chitin 1h | 8.38495 | 3.30225 | 1.19918 | 0.508494 |
| Epi 1h | 6.26419 | 3.27273 | 0.958673 | 0.507288 |
| SA 1h | 5.60596 | 3.25032 | 0.726526 | 0.421118 |
| Me-JA 1h | 5.80235 | 3.37108 | 0.943701 | 0.546579 |
| Control 6h | 12.607 | 6.46909 | 1.31166 | 0.685488 |
| ABA 6h | 54.8897 | 5.55523 | 6.79993 | 0.594246 |
| ACC 6h | 12.5259 | 4.21053 | 1.42523 | 0.528256 |
| BABA 6h | 15.2659 | 6.49203 | 1.50151 | 0.684341 |
| Chitin 6h | 9.38858 | 3.81365 | 1.11887 | 0.501291 |
| Epi 6h | 17.8907 | 4.08509 | 2.08807 | 0.501051 |
| SA 6h | 10.7659 | 3.88644 | 1.28508 | 0.552377 |
| Me-JA 6h | 19.5124 | 3.54679 | 2.55903 | 0.484064 |
Source Transcript PGSC0003DMT400009313 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G36250.1 | +1 | 2e-102 | 322 | 151/238 (63%) | aldehyde dehydrogenase 3F1 | chr4:17151029-17153381 FORWARD LENGTH=484 |
