Probe CUST_50228_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50228_PI426222305 | JHI_St_60k_v1 | DMT400080115 | GCTAAAACAATTCACGTTTCTCATGAATCAGTATTTCCTATAGATACTCGACTCACGTAA |
All Microarray Probes Designed to Gene DMG400031194
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50216_PI426222305 | JHI_St_60k_v1 | DMT400080118 | GCTAAAACAATTCACGTTTCTCATGAATCAGTATTTCCTATAGATACTCGACTCACGTAA |
| CUST_50228_PI426222305 | JHI_St_60k_v1 | DMT400080115 | GCTAAAACAATTCACGTTTCTCATGAATCAGTATTTCCTATAGATACTCGACTCACGTAA |
| CUST_50212_PI426222305 | JHI_St_60k_v1 | DMT400080117 | GCTAAAACAATTCACGTTTCTCATGAATCAGTATTTCCTATAGATACTCGACTCACGTAA |
| CUST_50241_PI426222305 | JHI_St_60k_v1 | DMT400080116 | GCTAAAACAATTCACGTTTCTCATGAATCAGTATTTCCTATAGATACTCGACTCACGTAA |
| CUST_50214_PI426222305 | JHI_St_60k_v1 | DMT400080119 | CTTCTGTTTCTGTTTTCCTGATTCAAACTGAGATCCATATGTCGATGAAAAGAACACATT |
| CUST_50222_PI426222305 | JHI_St_60k_v1 | DMT400080120 | GCTAAAACAATTCACGTTTCTCATGAATCAGTATTTCCTATAGATACTCGACTCACGTAA |
Microarray Signals from CUST_50228_PI426222305
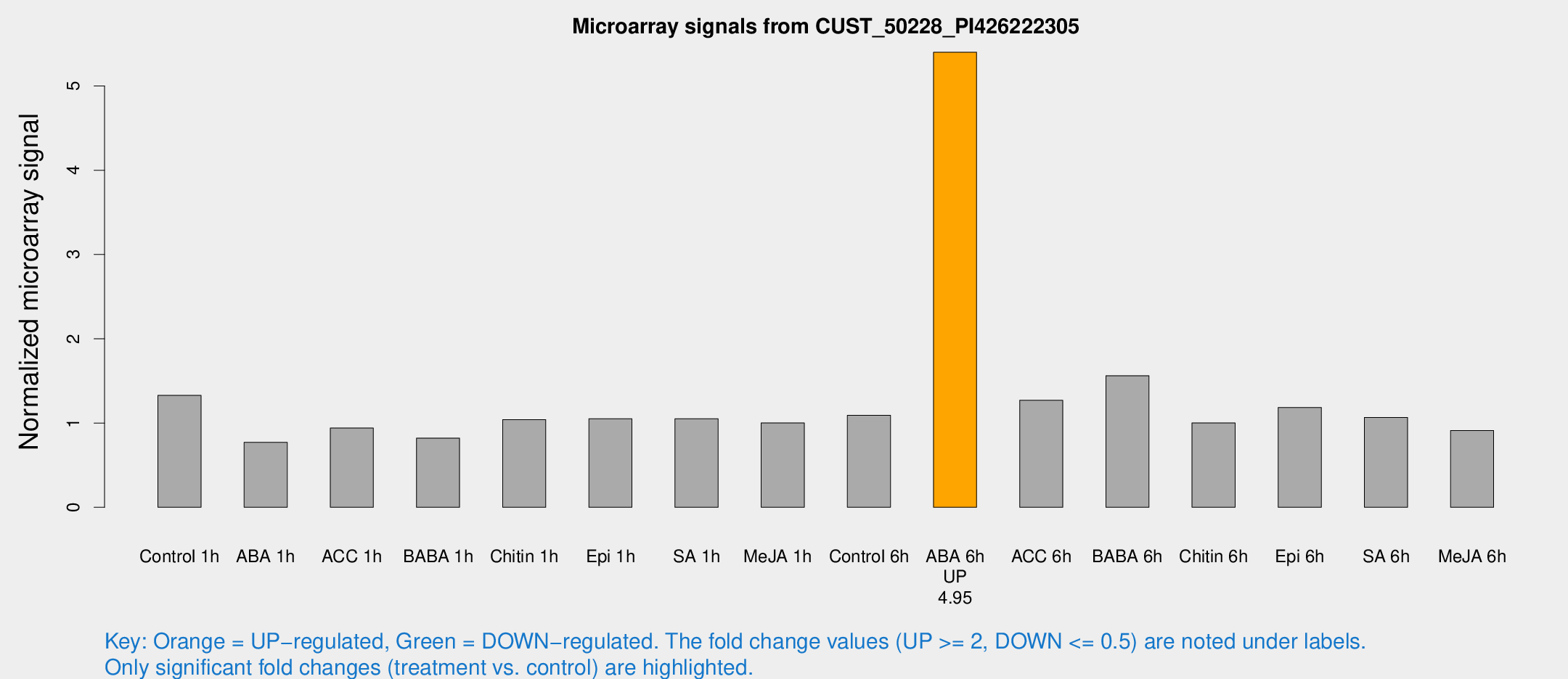
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 11.0797 | 3.33663 | 1.32756 | 0.455464 |
| ABA 1h | 5.38937 | 3.1281 | 0.770187 | 0.446669 |
| ACC 1h | 7.96598 | 3.54005 | 0.940048 | 0.460592 |
| BABA 1h | 6.274 | 3.49147 | 0.820739 | 0.458579 |
| Chitin 1h | 7.53251 | 3.31055 | 1.0405 | 0.473978 |
| Epi 1h | 7.88763 | 3.29449 | 1.05106 | 0.510728 |
| SA 1h | 9.39688 | 3.30629 | 1.051 | 0.448108 |
| Me-JA 1h | 6.58441 | 3.39175 | 1.00039 | 0.52681 |
| Control 6h | 8.8295 | 3.45835 | 1.09092 | 0.450568 |
| ABA 6h | 46.8441 | 8.2868 | 5.39986 | 0.679903 |
| ACC 6h | 12.8073 | 4.34993 | 1.26931 | 0.528444 |
| BABA 6h | 16.1718 | 5.52783 | 1.56176 | 0.6768 |
| Chitin 6h | 9.10616 | 3.79328 | 1.00041 | 0.483507 |
| Epi 6h | 10.6354 | 3.93958 | 1.1842 | 0.448793 |
| SA 6h | 8.80731 | 3.56736 | 1.06679 | 0.486768 |
| Me-JA 6h | 7.45737 | 3.38143 | 0.911742 | 0.450925 |
Source Transcript PGSC0003DMT400080115 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G13700.1 | +1 | 4e-121 | 384 | 178/249 (71%) | purple acid phosphatase 23 | chr4:7957072-7958919 REVERSE LENGTH=458 |
