Probe CUST_50130_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50130_PI426222305 | JHI_St_60k_v1 | DMT400065176 | CTCTATTTGAAGGTTAAAAGTGTCTTGACTTTGTAATTGAGATGCATTGTGTGAGCTATG |
All Microarray Probes Designed to Gene DMG400025331
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50130_PI426222305 | JHI_St_60k_v1 | DMT400065176 | CTCTATTTGAAGGTTAAAAGTGTCTTGACTTTGTAATTGAGATGCATTGTGTGAGCTATG |
| CUST_50107_PI426222305 | JHI_St_60k_v1 | DMT400065178 | TCCTACCGTGTATTCTCTATTTGAAGGTTAAAAGTGTCTTGACTTTGTAATTGAGATGCA |
| CUST_50117_PI426222305 | JHI_St_60k_v1 | DMT400065175 | AATATGGTCAACTCGACATAATGTTCAGTAACGCAGGAGTCGTTGGCCCTTTAGCTCATC |
Microarray Signals from CUST_50130_PI426222305
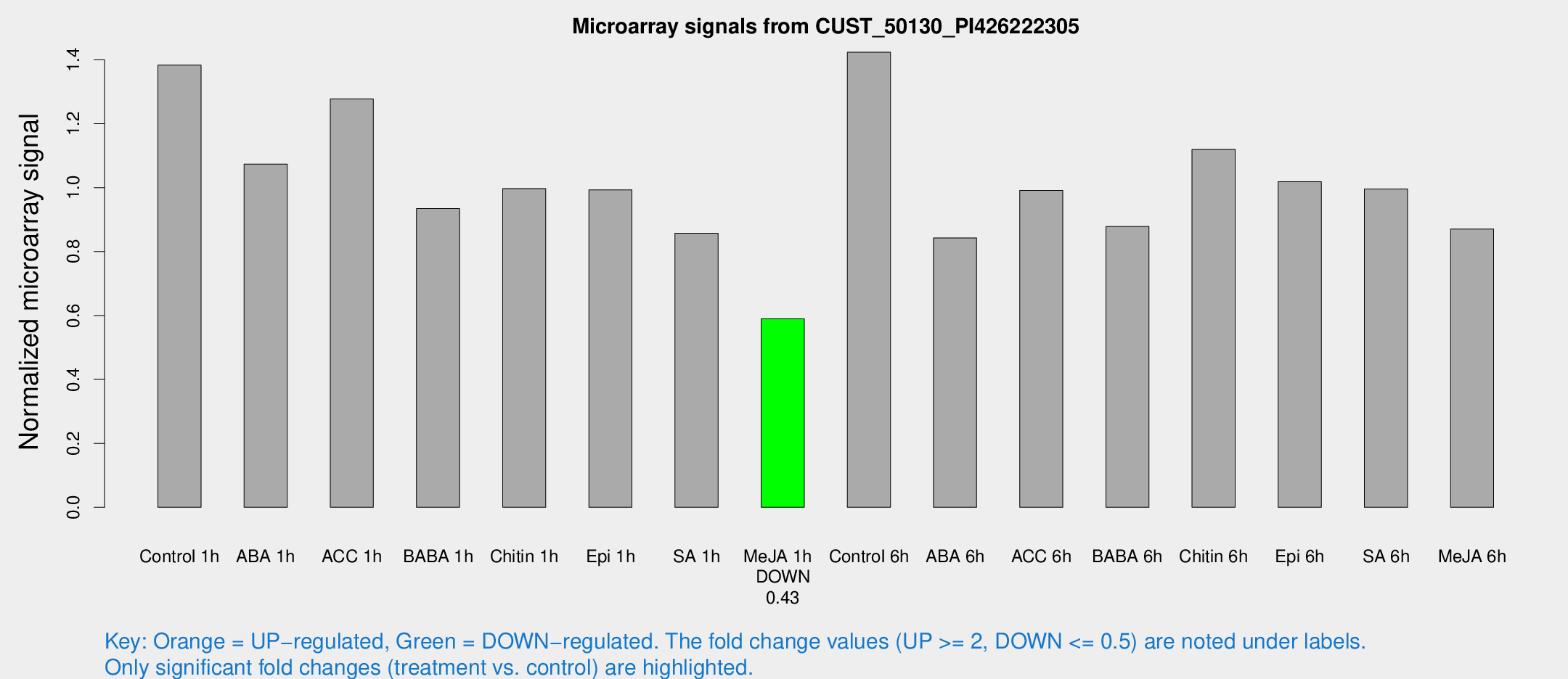
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1316.43 | 139.255 | 1.38317 | 0.0799249 |
| ABA 1h | 898.483 | 66.168 | 1.07371 | 0.11414 |
| ACC 1h | 1262.34 | 178.924 | 1.2775 | 0.0937081 |
| BABA 1h | 864.139 | 113.843 | 0.934475 | 0.0540779 |
| Chitin 1h | 855.145 | 94.3398 | 0.997169 | 0.0636941 |
| Epi 1h | 810.554 | 47.0025 | 0.992788 | 0.0574428 |
| SA 1h | 842.656 | 106.287 | 0.857269 | 0.0883855 |
| Me-JA 1h | 454.342 | 26.4861 | 0.589766 | 0.058566 |
| Control 6h | 1413.25 | 289.885 | 1.42356 | 0.202845 |
| ABA 6h | 849.917 | 74.358 | 0.842603 | 0.0816958 |
| ACC 6h | 1094.86 | 160.324 | 0.99122 | 0.0798608 |
| BABA 6h | 927.669 | 53.8122 | 0.878306 | 0.0508169 |
| Chitin 6h | 1125.29 | 65.2694 | 1.11971 | 0.064739 |
| Epi 6h | 1106.37 | 159.655 | 1.01875 | 0.24136 |
| SA 6h | 961.156 | 169.478 | 0.995617 | 0.0797733 |
| Me-JA 6h | 860.928 | 208.701 | 0.870631 | 0.155475 |
Source Transcript PGSC0003DMT400065176 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G47130.1 | +3 | 2e-80 | 252 | 135/250 (54%) | NAD(P)-binding Rossmann-fold superfamily protein | chr2:19349627-19350481 REVERSE LENGTH=257 |
