Probe CUST_50117_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50117_PI426222305 | JHI_St_60k_v1 | DMT400065175 | AATATGGTCAACTCGACATAATGTTCAGTAACGCAGGAGTCGTTGGCCCTTTAGCTCATC |
All Microarray Probes Designed to Gene DMG400025331
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50130_PI426222305 | JHI_St_60k_v1 | DMT400065176 | CTCTATTTGAAGGTTAAAAGTGTCTTGACTTTGTAATTGAGATGCATTGTGTGAGCTATG |
| CUST_50107_PI426222305 | JHI_St_60k_v1 | DMT400065178 | TCCTACCGTGTATTCTCTATTTGAAGGTTAAAAGTGTCTTGACTTTGTAATTGAGATGCA |
| CUST_50117_PI426222305 | JHI_St_60k_v1 | DMT400065175 | AATATGGTCAACTCGACATAATGTTCAGTAACGCAGGAGTCGTTGGCCCTTTAGCTCATC |
Microarray Signals from CUST_50117_PI426222305
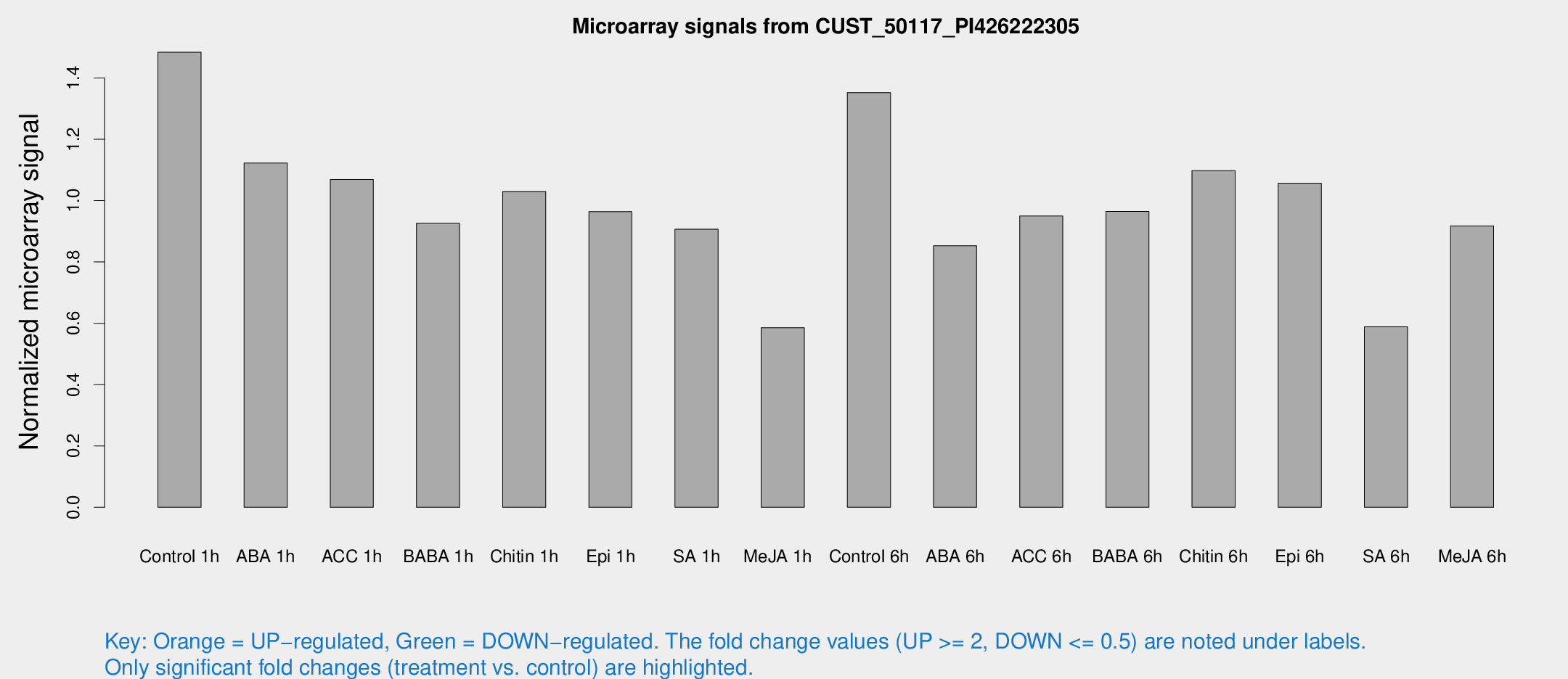
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 477.742 | 72.5405 | 1.48383 | 0.125932 |
| ABA 1h | 312.762 | 18.2937 | 1.12231 | 0.0655723 |
| ACC 1h | 383.191 | 113.073 | 1.06869 | 0.291353 |
| BABA 1h | 289.453 | 46.878 | 0.926054 | 0.0696068 |
| Chitin 1h | 298.014 | 42.9657 | 1.02951 | 0.0833345 |
| Epi 1h | 263.387 | 15.5227 | 0.96393 | 0.0566645 |
| SA 1h | 303.721 | 53.0089 | 0.906896 | 0.136919 |
| Me-JA 1h | 152.138 | 15.9915 | 0.585771 | 0.0616862 |
| Control 6h | 507.268 | 176.518 | 1.35218 | 0.545561 |
| ABA 6h | 285.759 | 16.8174 | 0.852647 | 0.0500966 |
| ACC 6h | 366.692 | 93.5251 | 0.94994 | 0.136259 |
| BABA 6h | 343.337 | 35.1229 | 0.964549 | 0.0796946 |
| Chitin 6h | 369.801 | 27.2553 | 1.09764 | 0.0641638 |
| Epi 6h | 384.654 | 63.7139 | 1.05679 | 0.190381 |
| SA 6h | 254.868 | 106.517 | 0.588618 | 0.408787 |
| Me-JA 6h | 301.901 | 69.7697 | 0.917244 | 0.135865 |
Source Transcript PGSC0003DMT400065175 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G47140.1 | +1 | 1e-29 | 108 | 53/94 (56%) | NAD(P)-binding Rossmann-fold superfamily protein | chr2:19350970-19352059 REVERSE LENGTH=257 |
