Probe CUST_48228_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48228_PI426222305 | JHI_St_60k_v1 | DMT400055911 | ACACGAAAAAGAAACCAACATAGTCAGGGATGAGGCAATTTTGTACTTCTTTTAGTTTCC |
All Microarray Probes Designed to Gene DMG400021719
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48216_PI426222305 | JHI_St_60k_v1 | DMT400055910 | GCAATGAATGTTCTTGGATTTGATATTAATGACACCAGTGCCACTGCCTCTAAAGGAGCT |
| CUST_48228_PI426222305 | JHI_St_60k_v1 | DMT400055911 | ACACGAAAAAGAAACCAACATAGTCAGGGATGAGGCAATTTTGTACTTCTTTTAGTTTCC |
Microarray Signals from CUST_48228_PI426222305
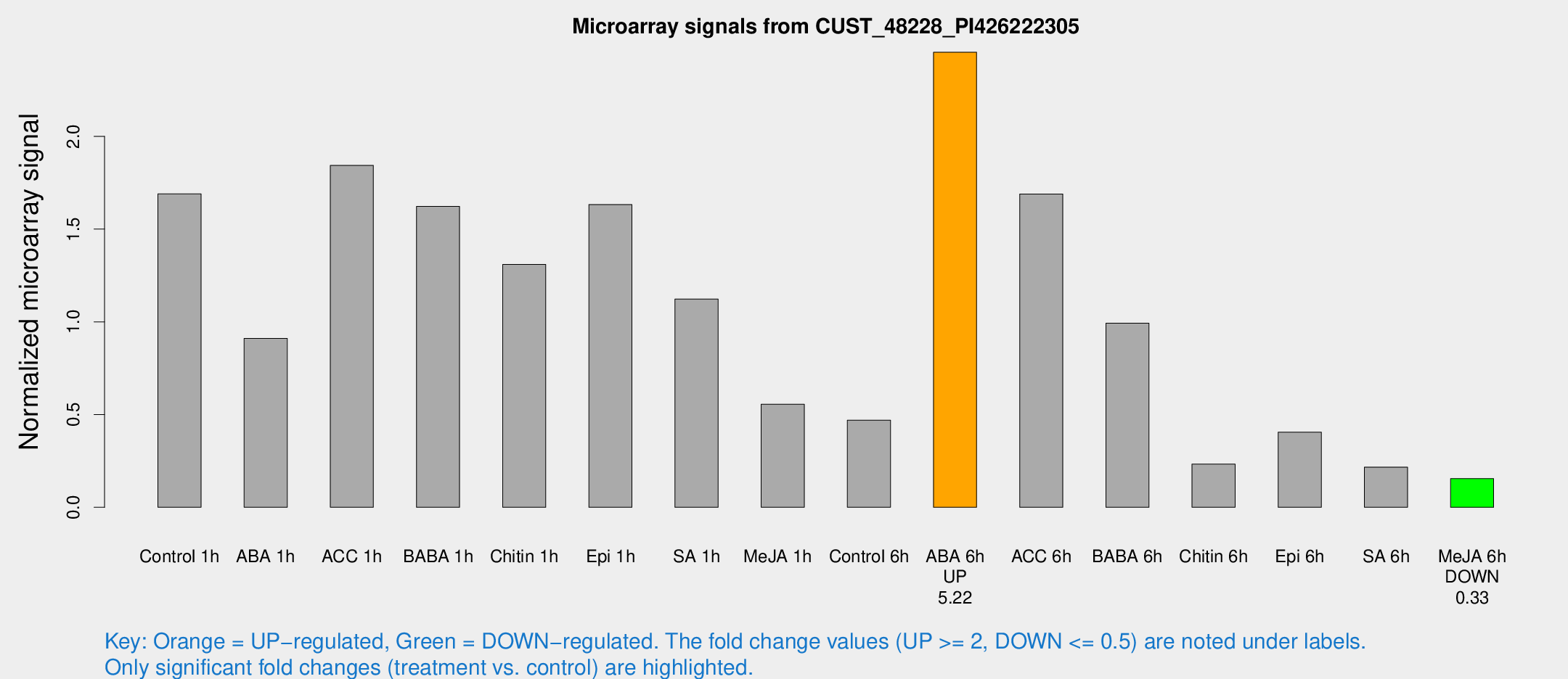
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 80.6696 | 15.2716 | 1.68962 | 0.39034 |
| ABA 1h | 38.0884 | 6.05101 | 0.910805 | 0.120424 |
| ACC 1h | 120.215 | 58.712 | 1.84343 | 1.242 |
| BABA 1h | 76.6099 | 19.1287 | 1.62239 | 0.614356 |
| Chitin 1h | 54.9008 | 5.63587 | 1.30963 | 0.195993 |
| Epi 1h | 65.5083 | 5.03424 | 1.63241 | 0.153652 |
| SA 1h | 69.4356 | 31.2505 | 1.12309 | 0.718147 |
| Me-JA 1h | 28.4587 | 15.1313 | 0.555892 | 0.513416 |
| Control 6h | 24.5378 | 8.39913 | 0.469791 | 0.143464 |
| ABA 6h | 124.667 | 24.6225 | 2.4538 | 0.355936 |
| ACC 6h | 109.97 | 52.3032 | 1.68945 | 1.14684 |
| BABA 6h | 51.2368 | 4.6689 | 0.992405 | 0.0904564 |
| Chitin 6h | 12.5828 | 3.59255 | 0.232772 | 0.0844546 |
| Epi 6h | 26.5684 | 10.3913 | 0.405054 | 0.314586 |
| SA 6h | 11.2951 | 4.22828 | 0.216763 | 0.122891 |
| Me-JA 6h | 7.57288 | 3.25077 | 0.154447 | 0.0751963 |
Source Transcript PGSC0003DMT400055911 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G09280.1 | +2 | 6e-09 | 51 | 26/56 (46%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: endomembrane system; EXPRESSED IN: root; Has 31 Blast hits to 31 proteins in 9 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 31; Viruses - 0; Other Eukaryotes - 0 (source: NCBI BLink). | chr3:2850920-2851258 REVERSE LENGTH=112 |
