Probe CUST_472_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_472_PI426222305 | JHI_St_60k_v1 | DMT400005471 | CAGTCCTGTTGTTGAAACTGATGATGTTGTGTACTCAATGTGAATACTAATAAGTAATCC |
All Microarray Probes Designed to Gene DMG400002134
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_472_PI426222305 | JHI_St_60k_v1 | DMT400005471 | CAGTCCTGTTGTTGAAACTGATGATGTTGTGTACTCAATGTGAATACTAATAAGTAATCC |
Microarray Signals from CUST_472_PI426222305
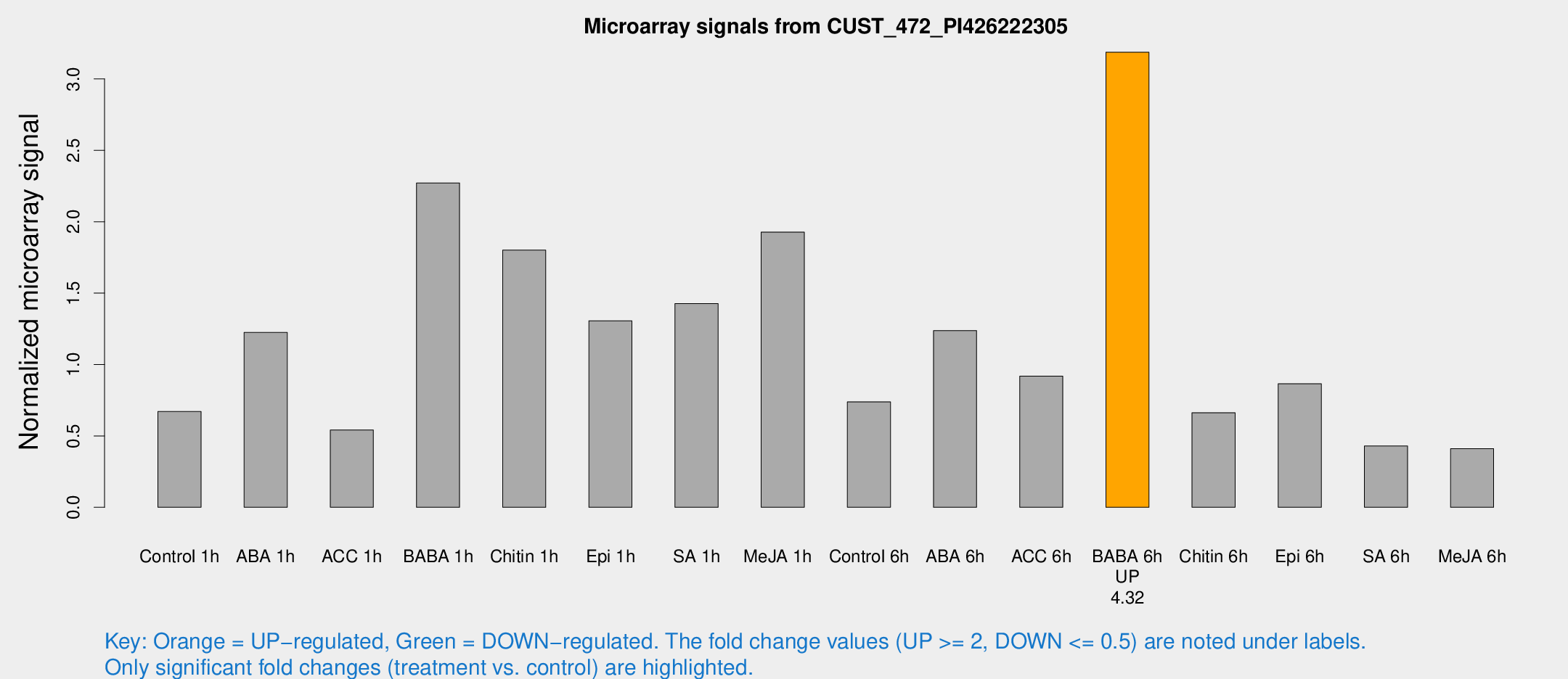
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 11.01 | 3.2817 | 0.670517 | 0.222788 |
| ABA 1h | 19.1658 | 5.86332 | 1.22475 | 0.451337 |
| ACC 1h | 9.28857 | 3.85827 | 0.541811 | 0.254236 |
| BABA 1h | 35.3959 | 5.73734 | 2.27171 | 0.274286 |
| Chitin 1h | 26.8089 | 5.34586 | 1.80111 | 0.585554 |
| Epi 1h | 20.8061 | 7.97113 | 1.30647 | 0.541174 |
| SA 1h | 24.2629 | 4.84093 | 1.42691 | 0.254227 |
| Me-JA 1h | 25.0071 | 3.60439 | 1.9274 | 0.343011 |
| Control 6h | 14.1176 | 6.08196 | 0.738568 | 0.287675 |
| ABA 6h | 25.6302 | 9.753 | 1.23759 | 0.734341 |
| ACC 6h | 21.4656 | 8.76464 | 0.918748 | 0.554278 |
| BABA 6h | 57.6117 | 8.84669 | 3.18724 | 0.461663 |
| Chitin 6h | 11.1958 | 3.70965 | 0.662422 | 0.223753 |
| Epi 6h | 15.9303 | 3.97398 | 0.864833 | 0.229596 |
| SA 6h | 6.76589 | 3.47138 | 0.429246 | 0.224429 |
| Me-JA 6h | 6.60543 | 3.30665 | 0.410997 | 0.213676 |
Source Transcript PGSC0003DMT400005471 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc04g074830.1 | +1 | 0.0 | 608 | 317/338 (94%) | evidence_code:10F0H1E1IEG genomic_reference:SL2.50ch04 gene_region:58341226-58342242 transcript_region:SL2.50ch04:58341226..58342242+ go_terms:GO:0005488 functional_description:At4g40080-like protein (Fragment) (AHRD V1 ***- B3SK93_ARALY); contains Interpro domain(s) IPR013809 Epsin-like, N-terminal |
| TAIR PP10 | AT4G40080.1 | +1 | 7e-48 | 171 | 114/292 (39%) | ENTH/ANTH/VHS superfamily protein | chr4:18579371-18580542 FORWARD LENGTH=365 |
