Probe CUST_47145_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47145_PI426222305 | JHI_St_60k_v1 | DMT400030404 | GCAGAGTTAGAGCTGGTAGAGGAATGGATTTATTTTGTACACATTTTTGTTTAGTAAGAG |
All Microarray Probes Designed to Gene DMG400011642
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47145_PI426222305 | JHI_St_60k_v1 | DMT400030404 | GCAGAGTTAGAGCTGGTAGAGGAATGGATTTATTTTGTACACATTTTTGTTTAGTAAGAG |
Microarray Signals from CUST_47145_PI426222305
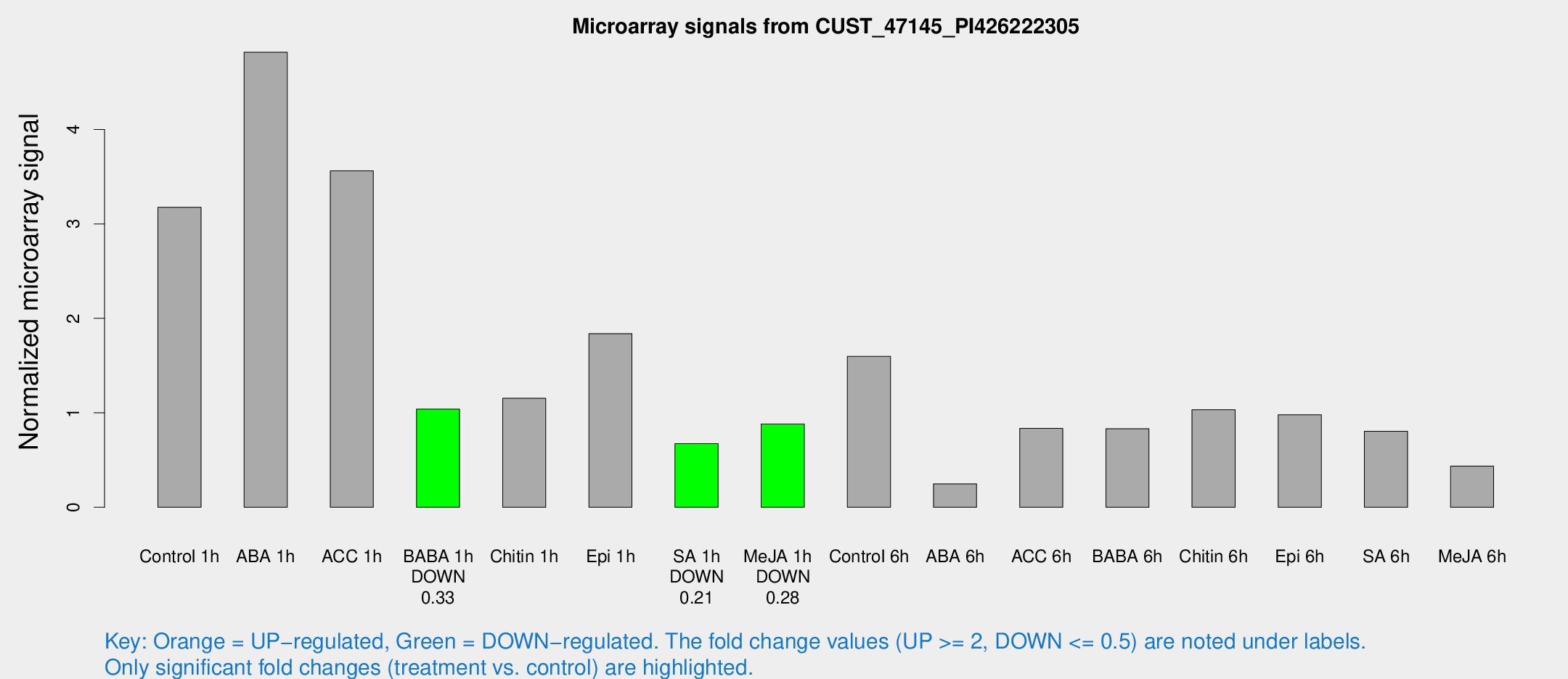
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 412.422 | 59.7079 | 3.17638 | 0.269781 |
| ABA 1h | 561.687 | 96.8167 | 4.81717 | 1.31437 |
| ACC 1h | 469.539 | 41.4281 | 3.56077 | 0.519968 |
| BABA 1h | 129.532 | 15.7852 | 1.03982 | 0.0696514 |
| Chitin 1h | 152.366 | 59.7301 | 1.15388 | 0.458541 |
| Epi 1h | 205.484 | 24.6883 | 1.83864 | 0.239699 |
| SA 1h | 89.5375 | 10.9918 | 0.674044 | 0.0468065 |
| Me-JA 1h | 91.9051 | 6.35696 | 0.881181 | 0.0790629 |
| Control 6h | 219.101 | 61.8345 | 1.59744 | 0.392982 |
| ABA 6h | 37.8144 | 12.1207 | 0.248167 | 0.116775 |
| ACC 6h | 128.669 | 29.7434 | 0.835638 | 0.086876 |
| BABA 6h | 131.491 | 38.2814 | 0.831816 | 0.331487 |
| Chitin 6h | 151.842 | 44.7824 | 1.03288 | 0.268231 |
| Epi 6h | 140.992 | 9.12207 | 0.979573 | 0.111798 |
| SA 6h | 111.949 | 30.1332 | 0.805248 | 0.180743 |
| Me-JA 6h | 57.344 | 11.2607 | 0.436534 | 0.0556879 |
Source Transcript PGSC0003DMT400030404 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G56360.1 | +3 | 5e-39 | 139 | 77/187 (41%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: chloroplast; EXPRESSED IN: 24 plant structures; EXPRESSED DURING: 15 growth stages; BEST Arabidopsis thaliana protein match is: unknown protein (TAIR:AT5G05250.1); Has 45 Blast hits to 45 proteins in 13 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 45; Viruses - 0; Other Eukaryotes - 0 (source: NCBI BLink). | chr3:20896641-20897342 FORWARD LENGTH=233 |
