Probe CUST_46857_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46857_PI426222305 | JHI_St_60k_v1 | DMT400081757 | TCAGTTGCATTTTTTGCTGAGGCTGGATTGGACTATGTTTCTTGCTCTCCATTCAGGGTG |
All Microarray Probes Designed to Gene DMG400032107
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46850_PI426222305 | JHI_St_60k_v1 | DMT400081755 | CAATGGGGGGTTTGCCAAGAACTTTTGAAAAAAGACGTTGTGTTTGAGATTAATTGTATT |
| CUST_46857_PI426222305 | JHI_St_60k_v1 | DMT400081757 | TCAGTTGCATTTTTTGCTGAGGCTGGATTGGACTATGTTTCTTGCTCTCCATTCAGGGTG |
| CUST_46846_PI426222305 | JHI_St_60k_v1 | DMT400081758 | ATATACCTAGCGAAAGGCATCCTCCAACACGATCCATTCGAGGTTCTTGATCAAAAGGGT |
Microarray Signals from CUST_46857_PI426222305
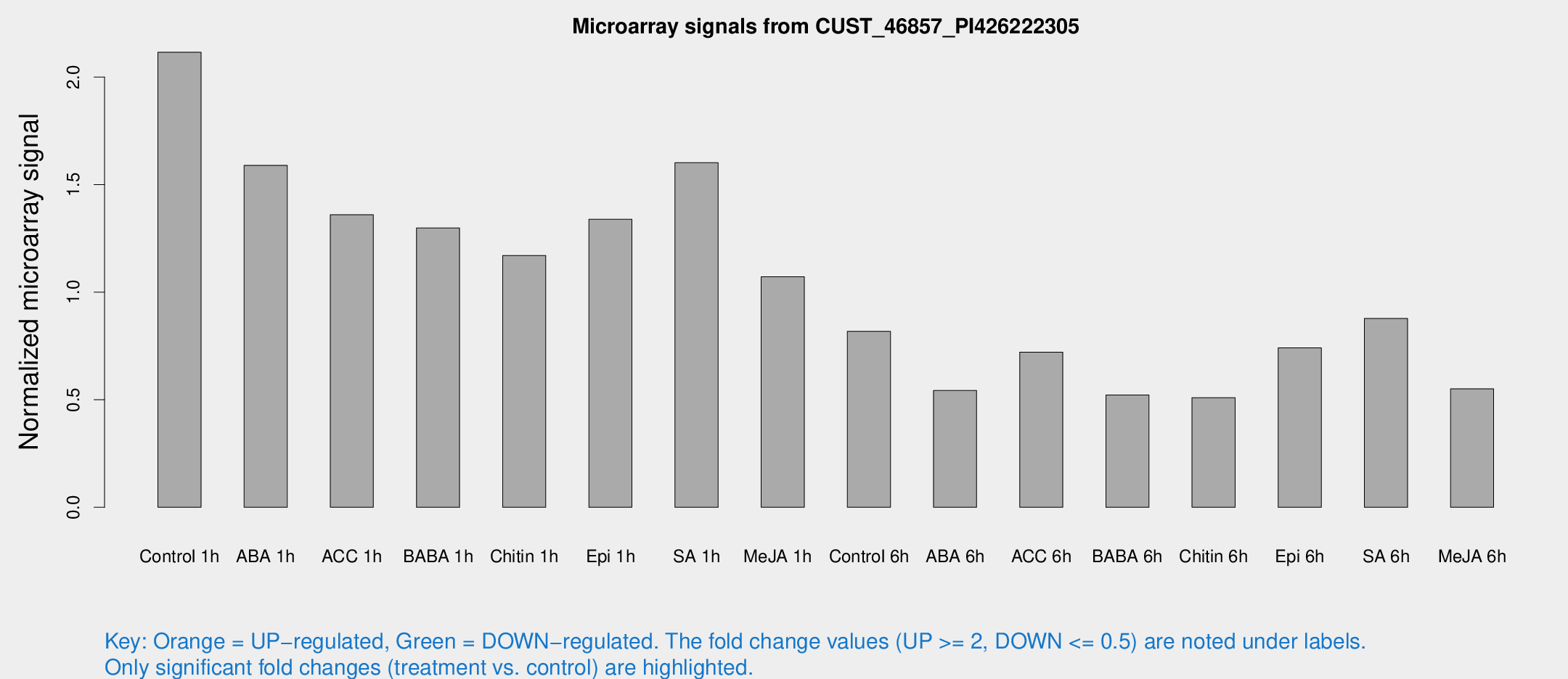
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 29634.8 | 2114.53 | 2.11549 | 0.122138 |
| ABA 1h | 19664.5 | 1168.18 | 1.58947 | 0.0917686 |
| ACC 1h | 20630.2 | 4685.19 | 1.36 | 0.250568 |
| BABA 1h | 19655.4 | 6058.73 | 1.29839 | 0.361203 |
| Chitin 1h | 14805.6 | 1445.93 | 1.17076 | 0.0675943 |
| Epi 1h | 16194 | 937.691 | 1.33864 | 0.081677 |
| SA 1h | 23729.7 | 4037.59 | 1.60228 | 0.222649 |
| Me-JA 1h | 12300.1 | 1086.27 | 1.07212 | 0.0690669 |
| Control 6h | 11879 | 2389.54 | 0.818242 | 0.126433 |
| ABA 6h | 8311.21 | 1390.58 | 0.542902 | 0.0784338 |
| ACC 6h | 11567.6 | 669.674 | 0.721468 | 0.0795231 |
| BABA 6h | 8227.14 | 871.563 | 0.521713 | 0.0491187 |
| Chitin 6h | 7570.07 | 437.708 | 0.509515 | 0.0294181 |
| Epi 6h | 12266.6 | 2564.02 | 0.741591 | 0.135717 |
| SA 6h | 12148.5 | 762.4 | 0.877629 | 0.153031 |
| Me-JA 6h | 7730.22 | 816.966 | 0.550768 | 0.0317998 |
Source Transcript PGSC0003DMT400081757 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G15530.5 | +3 | 0.0 | 543 | 271/342 (79%) | pyruvate orthophosphate dikinase | chr4:8864828-8870748 REVERSE LENGTH=963 |
