Probe CUST_45552_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45552_PI426222305 | JHI_St_60k_v1 | DMT400079707 | CCCTCTCTGCGTCATCTTTGAATTTCACGTGCAATAATATTTGAAATCTACACCTATTAT |
All Microarray Probes Designed to Gene DMG400031042
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45552_PI426222305 | JHI_St_60k_v1 | DMT400079707 | CCCTCTCTGCGTCATCTTTGAATTTCACGTGCAATAATATTTGAAATCTACACCTATTAT |
| CUST_45524_PI426222305 | JHI_St_60k_v1 | DMT400079708 | CCCTCTCTGCGTCATCTTTGAATTTCACGTGCAATAATATTTGAAATCTACACCTATTAT |
| CUST_45526_PI426222305 | JHI_St_60k_v1 | DMT400079706 | TATTATCAGTTAATAAGGCTTGGCTTTGAGGGTTATAAGGACGTCATGGAGAATTGCTTA |
Microarray Signals from CUST_45552_PI426222305
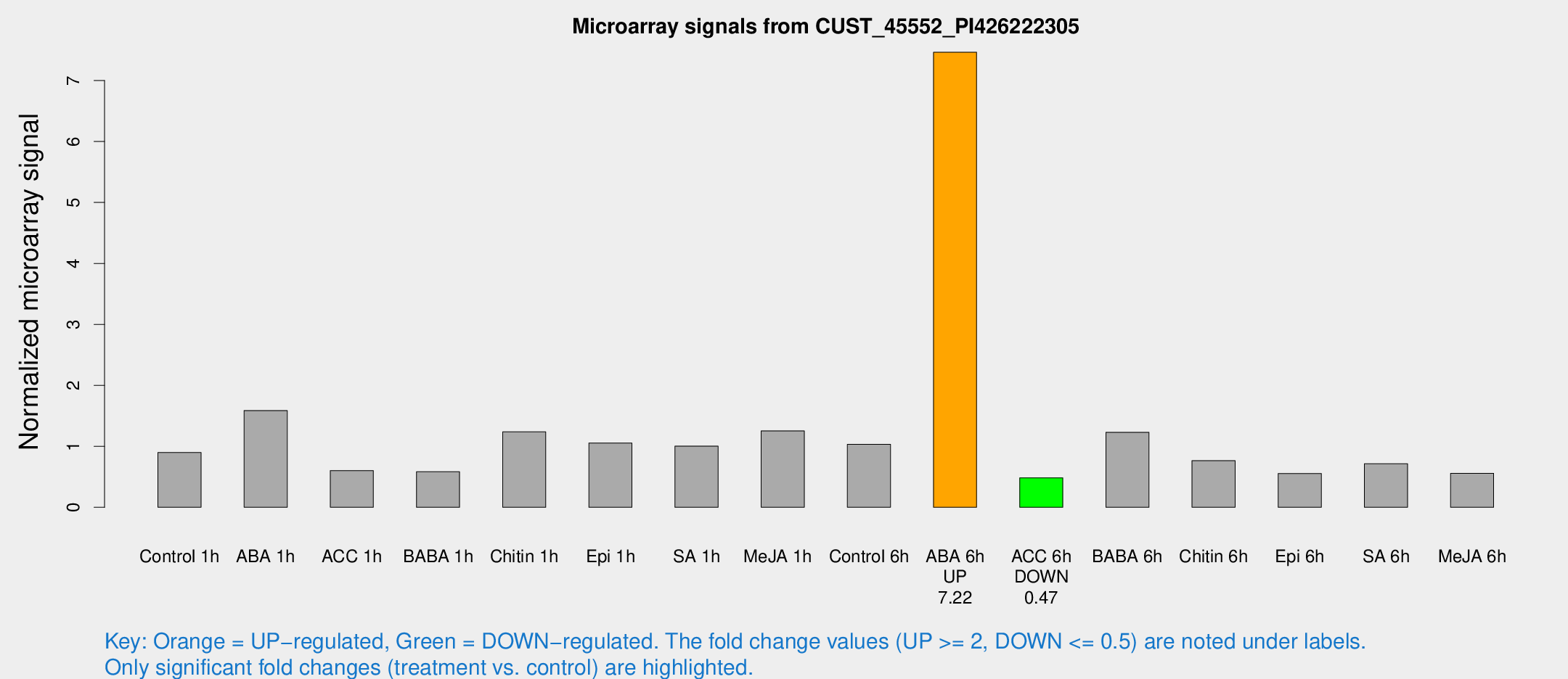
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 17.6625 | 3.23046 | 0.898948 | 0.171943 |
| ABA 1h | 29.0598 | 7.2472 | 1.58776 | 0.503823 |
| ACC 1h | 13.5437 | 4.4636 | 0.603053 | 0.210322 |
| BABA 1h | 12.2264 | 3.85843 | 0.58311 | 0.302779 |
| Chitin 1h | 23.3419 | 6.89432 | 1.23708 | 0.258022 |
| Epi 1h | 18.0595 | 3.26758 | 1.05456 | 0.198131 |
| SA 1h | 22.9661 | 8.38719 | 1.00377 | 0.469555 |
| Me-JA 1h | 22.0731 | 6.60568 | 1.25453 | 0.62011 |
| Control 6h | 21.1862 | 4.45226 | 1.0339 | 0.203972 |
| ABA 6h | 172.124 | 52.5084 | 7.46432 | 3.09165 |
| ACC 6h | 11.0159 | 4.01452 | 0.483663 | 0.173067 |
| BABA 6h | 26.8337 | 4.0352 | 1.23063 | 0.18779 |
| Chitin 6h | 16.2052 | 3.73739 | 0.764808 | 0.186737 |
| Epi 6h | 14.1519 | 5.91132 | 0.55276 | 0.29992 |
| SA 6h | 14.4538 | 3.57663 | 0.714822 | 0.269771 |
| Me-JA 6h | 13.5605 | 6.72331 | 0.557076 | 0.265651 |
Source Transcript PGSC0003DMT400079707 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G17330.1 | +3 | 0.0 | 557 | 288/333 (86%) | glutamate decarboxylase | chr5:5711141-5714839 FORWARD LENGTH=502 |
