Probe CUST_45524_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45524_PI426222305 | JHI_St_60k_v1 | DMT400079708 | CCCTCTCTGCGTCATCTTTGAATTTCACGTGCAATAATATTTGAAATCTACACCTATTAT |
All Microarray Probes Designed to Gene DMG400031042
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45552_PI426222305 | JHI_St_60k_v1 | DMT400079707 | CCCTCTCTGCGTCATCTTTGAATTTCACGTGCAATAATATTTGAAATCTACACCTATTAT |
| CUST_45524_PI426222305 | JHI_St_60k_v1 | DMT400079708 | CCCTCTCTGCGTCATCTTTGAATTTCACGTGCAATAATATTTGAAATCTACACCTATTAT |
| CUST_45526_PI426222305 | JHI_St_60k_v1 | DMT400079706 | TATTATCAGTTAATAAGGCTTGGCTTTGAGGGTTATAAGGACGTCATGGAGAATTGCTTA |
Microarray Signals from CUST_45524_PI426222305
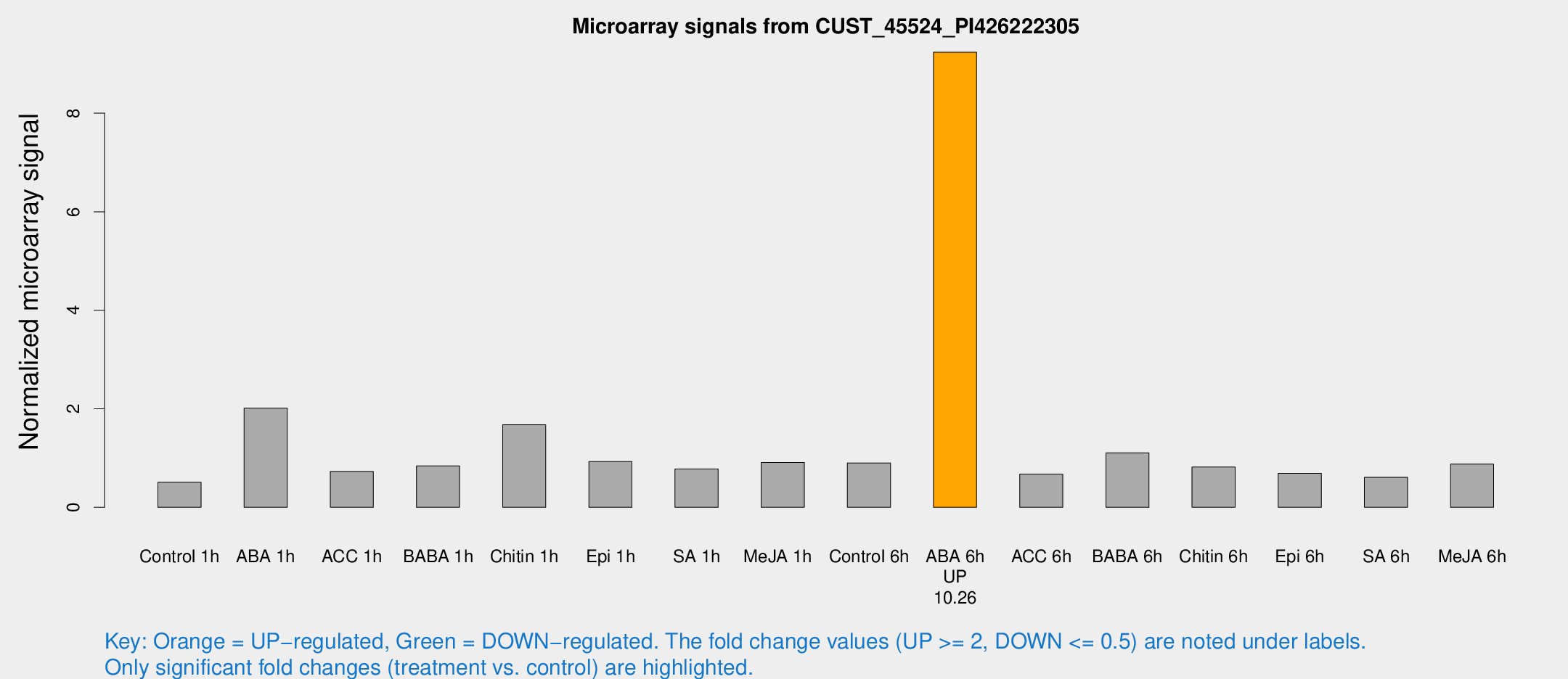
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 9.6273 | 3.09257 | 0.509428 | 0.17157 |
| ABA 1h | 33.4324 | 3.92838 | 2.01582 | 0.219112 |
| ACC 1h | 16.1059 | 5.32284 | 0.727796 | 0.274541 |
| BABA 1h | 15.2989 | 3.35495 | 0.84037 | 0.219731 |
| Chitin 1h | 28.7431 | 5.31749 | 1.67552 | 0.213553 |
| Epi 1h | 18.1868 | 6.62192 | 0.92897 | 0.52939 |
| SA 1h | 18.828 | 7.49138 | 0.779471 | 0.471928 |
| Me-JA 1h | 15.8755 | 5.23175 | 0.90877 | 0.546851 |
| Control 6h | 19.4328 | 6.24432 | 0.900803 | 0.327256 |
| ABA 6h | 187.489 | 33.5664 | 9.23884 | 2.0279 |
| ACC 6h | 15.1518 | 3.9973 | 0.6734 | 0.29759 |
| BABA 6h | 23.6095 | 3.91205 | 1.10633 | 0.229025 |
| Chitin 6h | 19.6204 | 6.98405 | 0.817752 | 0.457009 |
| Epi 6h | 14.6684 | 3.80379 | 0.689034 | 0.189413 |
| SA 6h | 12.2439 | 3.94474 | 0.609765 | 0.211192 |
| Me-JA 6h | 17.4915 | 5.25017 | 0.875801 | 0.201346 |
Source Transcript PGSC0003DMT400079708 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G17330.1 | +2 | 1e-50 | 181 | 95/174 (55%) | glutamate decarboxylase | chr5:5711141-5714839 FORWARD LENGTH=502 |
