Probe CUST_45377_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45377_PI426222305 | JHI_St_60k_v1 | DMT400001604 | ACATTGGTGATGTCATTCAGGTGCTAAGCAACAATAAATTCAAGAGTGCAACTCACAAAG |
All Microarray Probes Designed to Gene DMG404000594
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45357_PI426222305 | JHI_St_60k_v1 | DMT400001608 | GAAAAACTATCGGTACCTTCTGATACTTATTTCAAAAGCACAAAGAAATCAGCTGGGACT |
| CUST_45366_PI426222305 | JHI_St_60k_v1 | DMT400001603 | CAAGCCTGATTCTTTGAAGAGGAATCCTCAATGCACTGTTTGTTGTTACTGATTTAAAAT |
| CUST_45377_PI426222305 | JHI_St_60k_v1 | DMT400001604 | ACATTGGTGATGTCATTCAGGTGCTAAGCAACAATAAATTCAAGAGTGCAACTCACAAAG |
| CUST_45344_PI426222305 | JHI_St_60k_v1 | DMT400001605 | GCCTTCCAAAAACAAACTAGTTGTTAACATTGGTGATGTCATTCAGGTATTTATAACTCC |
| CUST_45365_PI426222305 | JHI_St_60k_v1 | DMT400001607 | CGCCTTCCAAACACAAACTAATTGTTAACATTGGTGATGTCATTCAGGTGCTAAGCAACA |
Microarray Signals from CUST_45377_PI426222305
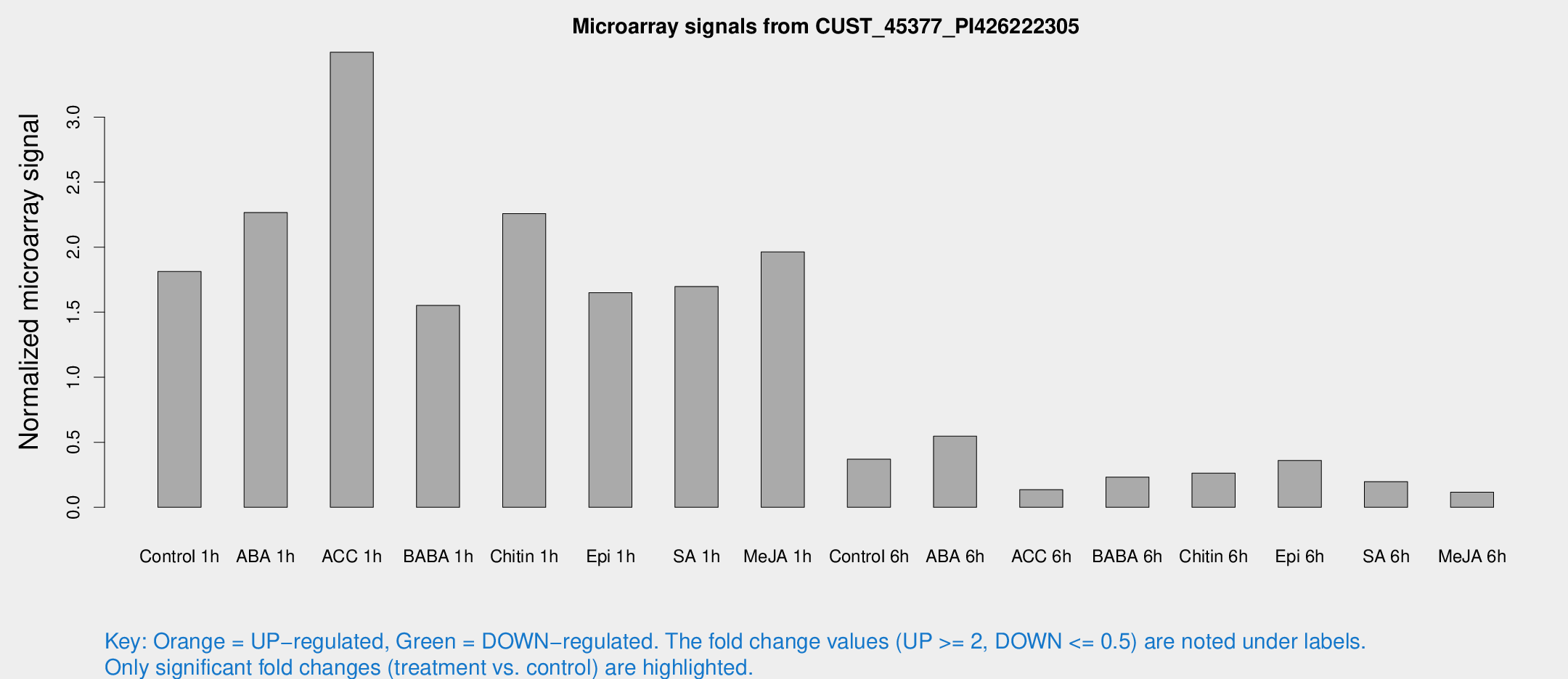
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 867.955 | 222.62 | 1.81308 | 0.331135 |
| ABA 1h | 972.374 | 232.758 | 2.26655 | 0.714631 |
| ACC 1h | 1791.75 | 499.188 | 3.49844 | 1.53363 |
| BABA 1h | 679.814 | 62.8107 | 1.55177 | 0.268007 |
| Chitin 1h | 963.816 | 225.974 | 2.25824 | 0.626471 |
| Epi 1h | 656.604 | 98.4086 | 1.64959 | 0.194317 |
| SA 1h | 821.471 | 181.226 | 1.69785 | 0.46347 |
| Me-JA 1h | 741.08 | 113.416 | 1.96381 | 0.485615 |
| Control 6h | 224.396 | 95.5005 | 0.370182 | 0.235365 |
| ABA 6h | 271.201 | 51.8369 | 0.546959 | 0.126908 |
| ACC 6h | 74.6445 | 20.0158 | 0.135035 | 0.0164096 |
| BABA 6h | 118.259 | 11.9387 | 0.232317 | 0.0223852 |
| Chitin 6h | 132.099 | 27.3356 | 0.262597 | 0.0560788 |
| Epi 6h | 188.958 | 32.8384 | 0.359703 | 0.0922796 |
| SA 6h | 91.783 | 18.9394 | 0.19665 | 0.0236651 |
| Me-JA 6h | 59.6616 | 22.982 | 0.116196 | 0.0351047 |
Source Transcript PGSC0003DMT400001604 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G10490.1 | +1 | 6e-37 | 139 | 89/271 (33%) | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | chr4:6483900-6485179 FORWARD LENGTH=348 |
