Probe CUST_33958_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33958_PI426222305 | JHI_St_60k_v1 | DMT400012345 | TGCAGGGTATGATAGTAGAAAGAATTCAAGCCAGCATTGCTAAGGAAGCGAAGAGAAGAA |
All Microarray Probes Designed to Gene DMG402004840
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33958_PI426222305 | JHI_St_60k_v1 | DMT400012345 | TGCAGGGTATGATAGTAGAAAGAATTCAAGCCAGCATTGCTAAGGAAGCGAAGAGAAGAA |
| CUST_33857_PI426222305 | JHI_St_60k_v1 | DMT400012353 | TATGTTGATTTTCAAACATCCCCTCATGGTTGCAATCTCTGCCCTCCCAACATGTGCAAG |
Microarray Signals from CUST_33958_PI426222305
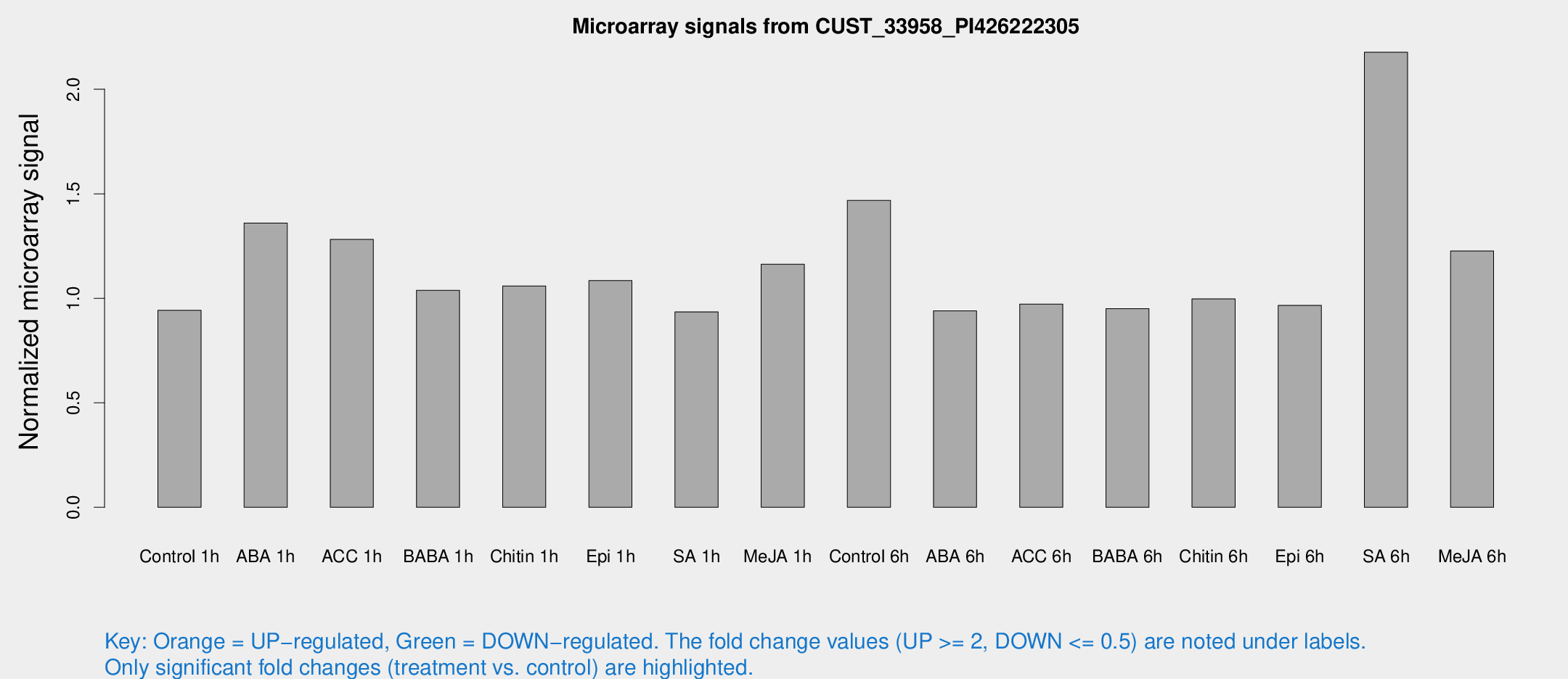
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.38932 | 3.12256 | 0.94296 | 0.546023 |
| ABA 1h | 7.43513 | 2.98793 | 1.35976 | 0.647726 |
| ACC 1h | 7.98098 | 3.44703 | 1.28179 | 0.624155 |
| BABA 1h | 5.72405 | 3.30785 | 1.03839 | 0.599672 |
| Chitin 1h | 5.35457 | 3.11741 | 1.05909 | 0.602331 |
| Epi 1h | 5.32504 | 3.09437 | 1.08463 | 0.623261 |
| SA 1h | 5.49262 | 3.09745 | 0.934868 | 0.528078 |
| Me-JA 1h | 5.44468 | 3.16461 | 1.16327 | 0.673552 |
| Control 6h | 9.97114 | 4.32395 | 1.46817 | 0.677966 |
| ABA 6h | 5.71277 | 3.31236 | 0.93986 | 0.544434 |
| ACC 6h | 6.53044 | 3.88009 | 0.972582 | 0.563194 |
| BABA 6h | 6.08842 | 3.53142 | 0.950742 | 0.550581 |
| Chitin 6h | 6.06682 | 3.51782 | 0.997289 | 0.577948 |
| Epi 6h | 6.28081 | 3.67429 | 0.966458 | 0.560396 |
| SA 6h | 14.6537 | 5.80738 | 2.17745 | 1.46534 |
| Me-JA 6h | 7.50489 | 3.17165 | 1.22626 | 0.595626 |
Source Transcript PGSC0003DMT400012345 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G73010.1 | +1 | 5e-36 | 129 | 67/128 (52%) | phosphate starvation-induced gene 2 | chr1:27464780-27466180 REVERSE LENGTH=295 |
