Probe CUST_33857_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33857_PI426222305 | JHI_St_60k_v1 | DMT400012353 | TATGTTGATTTTCAAACATCCCCTCATGGTTGCAATCTCTGCCCTCCCAACATGTGCAAG |
All Microarray Probes Designed to Gene DMG402004840
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33958_PI426222305 | JHI_St_60k_v1 | DMT400012345 | TGCAGGGTATGATAGTAGAAAGAATTCAAGCCAGCATTGCTAAGGAAGCGAAGAGAAGAA |
| CUST_33857_PI426222305 | JHI_St_60k_v1 | DMT400012353 | TATGTTGATTTTCAAACATCCCCTCATGGTTGCAATCTCTGCCCTCCCAACATGTGCAAG |
Microarray Signals from CUST_33857_PI426222305
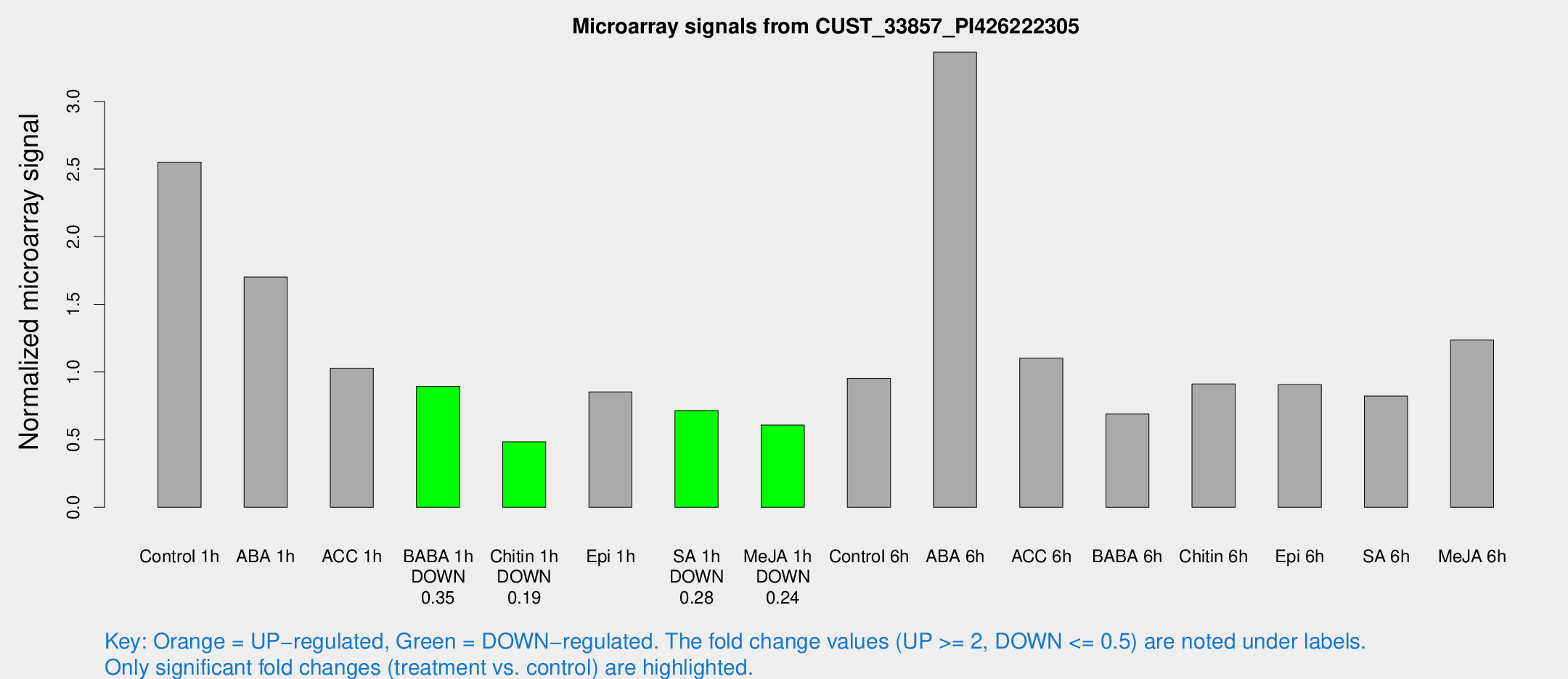
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 227.547 | 43.291 | 2.5507 | 0.333745 |
| ABA 1h | 139.712 | 33.8703 | 1.70189 | 0.422772 |
| ACC 1h | 103.164 | 31.9046 | 1.02845 | 0.324875 |
| BABA 1h | 79.3418 | 19.1635 | 0.894963 | 0.152613 |
| Chitin 1h | 37.7739 | 3.87068 | 0.483728 | 0.0495493 |
| Epi 1h | 69.5423 | 17.9228 | 0.852385 | 0.276276 |
| SA 1h | 68.118 | 16.2216 | 0.715378 | 0.167566 |
| Me-JA 1h | 43.2778 | 4.59184 | 0.607643 | 0.0580606 |
| Control 6h | 93.4212 | 34.6772 | 0.953926 | 0.333184 |
| ABA 6h | 327.164 | 71.9714 | 3.3635 | 0.704891 |
| ACC 6h | 111.985 | 18.6329 | 1.10205 | 0.0753789 |
| BABA 6h | 69.2515 | 13.9776 | 0.68972 | 0.125458 |
| Chitin 6h | 86.7408 | 15.0548 | 0.911677 | 0.186484 |
| Epi 6h | 89.2374 | 9.76291 | 0.90656 | 0.065281 |
| SA 6h | 70.1596 | 5.41681 | 0.821771 | 0.102057 |
| Me-JA 6h | 110.445 | 22.8368 | 1.23586 | 0.158659 |
Source Transcript PGSC0003DMT400012353 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G73010.1 | +1 | 5e-128 | 372 | 181/278 (65%) | phosphate starvation-induced gene 2 | chr1:27464780-27466180 REVERSE LENGTH=295 |
