Probe CUST_32315_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32315_PI426222305 | JHI_St_60k_v1 | DMT400012612 | TCAGAGAGCTCTCTGGAAGAGGACAAGACATTTACATACGAATGGATTCTTCAGACATAG |
All Microarray Probes Designed to Gene DMG400004930
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32315_PI426222305 | JHI_St_60k_v1 | DMT400012612 | TCAGAGAGCTCTCTGGAAGAGGACAAGACATTTACATACGAATGGATTCTTCAGACATAG |
| CUST_32325_PI426222305 | JHI_St_60k_v1 | DMT400012611 | TCAGAGAGCTCTCTGGAAGAGGACAAGACATTTACATACGAATGGATTCTTCAGACATAG |
| CUST_32328_PI426222305 | JHI_St_60k_v1 | DMT400012609 | TCAGAGAGCTCTCTGGAAGAGGACAAGACATTTACATACGAATGGATTCTTCAGACATAG |
| CUST_32249_PI426222305 | JHI_St_60k_v1 | DMT400012608 | TCAGAGAGCTCTCTGGAAGAGGACAAGACATTTACATACGAATGGATTCTTCAGACATAG |
| CUST_32219_PI426222305 | JHI_St_60k_v1 | DMT400012610 | TCAGAGAGCTCTCTGGAAGAGGACAAGACATTTACATACGAATGGATTCTTCAGACATAG |
Microarray Signals from CUST_32315_PI426222305
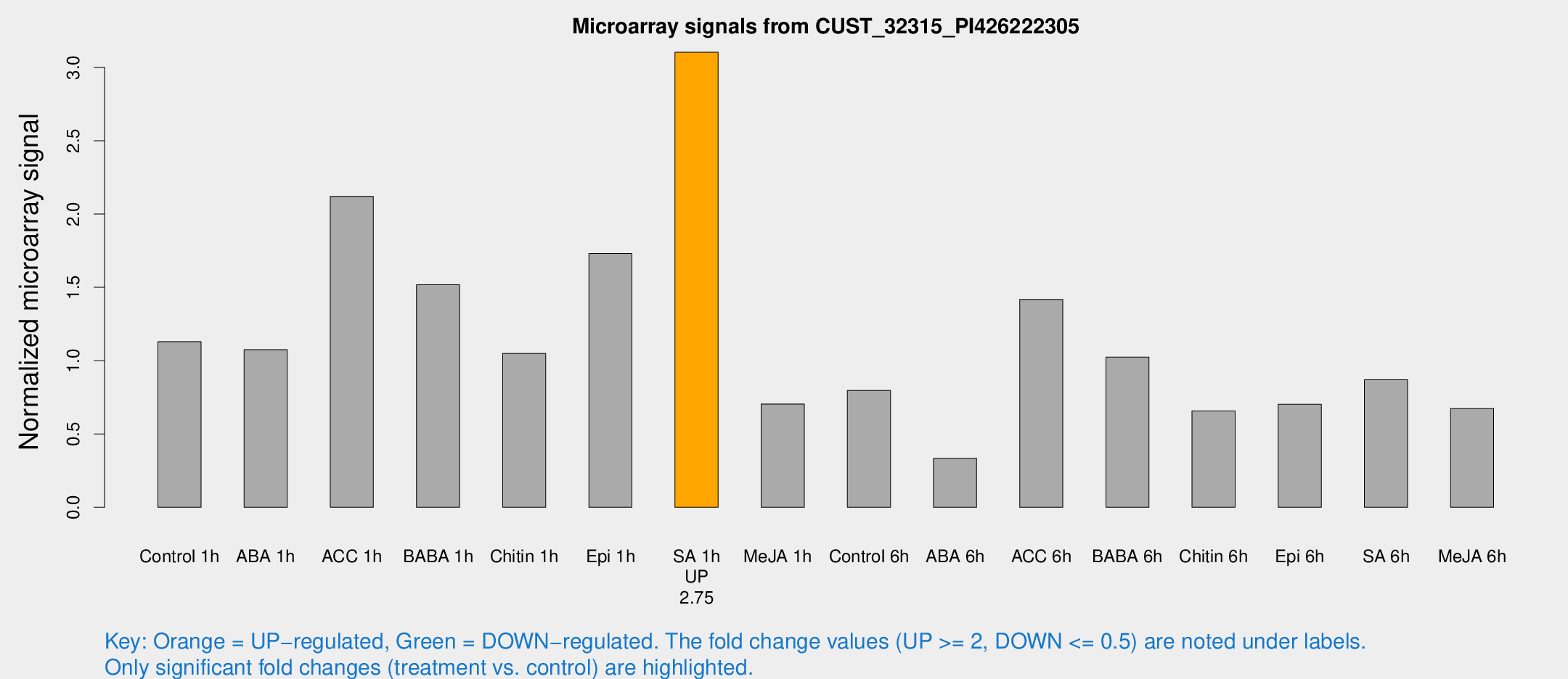
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 149.351 | 22.8471 | 1.12969 | 0.0961303 |
| ABA 1h | 123.795 | 10.4952 | 1.07513 | 0.171696 |
| ACC 1h | 338.354 | 121.636 | 2.12074 | 0.924619 |
| BABA 1h | 204.732 | 54.3445 | 1.51769 | 0.304874 |
| Chitin 1h | 127.918 | 27.69 | 1.04882 | 0.213168 |
| Epi 1h | 193.607 | 11.6767 | 1.73053 | 0.104301 |
| SA 1h | 436.252 | 94.3903 | 3.10336 | 0.621969 |
| Me-JA 1h | 74.9686 | 7.07997 | 0.704628 | 0.0525011 |
| Control 6h | 110.97 | 26.3346 | 0.797031 | 0.156323 |
| ABA 6h | 53.2829 | 19.6295 | 0.334299 | 0.123956 |
| ACC 6h | 211.208 | 15.5547 | 1.41755 | 0.23722 |
| BABA 6h | 160.61 | 46.7189 | 1.02434 | 0.260638 |
| Chitin 6h | 98.5295 | 25.3569 | 0.657093 | 0.218754 |
| Epi 6h | 115.465 | 36.307 | 0.702768 | 0.235751 |
| SA 6h | 115.69 | 23.7194 | 0.870012 | 0.294874 |
| Me-JA 6h | 93.2028 | 23.9932 | 0.67328 | 0.132728 |
Source Transcript PGSC0003DMT400012612 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G27290.1 | +3 | 0.0 | 847 | 425/789 (54%) | S-locus lectin protein kinase family protein | chr4:13666281-13669202 FORWARD LENGTH=783 |
