Probe CUST_32219_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32219_PI426222305 | JHI_St_60k_v1 | DMT400012610 | TCAGAGAGCTCTCTGGAAGAGGACAAGACATTTACATACGAATGGATTCTTCAGACATAG |
All Microarray Probes Designed to Gene DMG400004930
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32315_PI426222305 | JHI_St_60k_v1 | DMT400012612 | TCAGAGAGCTCTCTGGAAGAGGACAAGACATTTACATACGAATGGATTCTTCAGACATAG |
| CUST_32325_PI426222305 | JHI_St_60k_v1 | DMT400012611 | TCAGAGAGCTCTCTGGAAGAGGACAAGACATTTACATACGAATGGATTCTTCAGACATAG |
| CUST_32328_PI426222305 | JHI_St_60k_v1 | DMT400012609 | TCAGAGAGCTCTCTGGAAGAGGACAAGACATTTACATACGAATGGATTCTTCAGACATAG |
| CUST_32249_PI426222305 | JHI_St_60k_v1 | DMT400012608 | TCAGAGAGCTCTCTGGAAGAGGACAAGACATTTACATACGAATGGATTCTTCAGACATAG |
| CUST_32219_PI426222305 | JHI_St_60k_v1 | DMT400012610 | TCAGAGAGCTCTCTGGAAGAGGACAAGACATTTACATACGAATGGATTCTTCAGACATAG |
Microarray Signals from CUST_32219_PI426222305
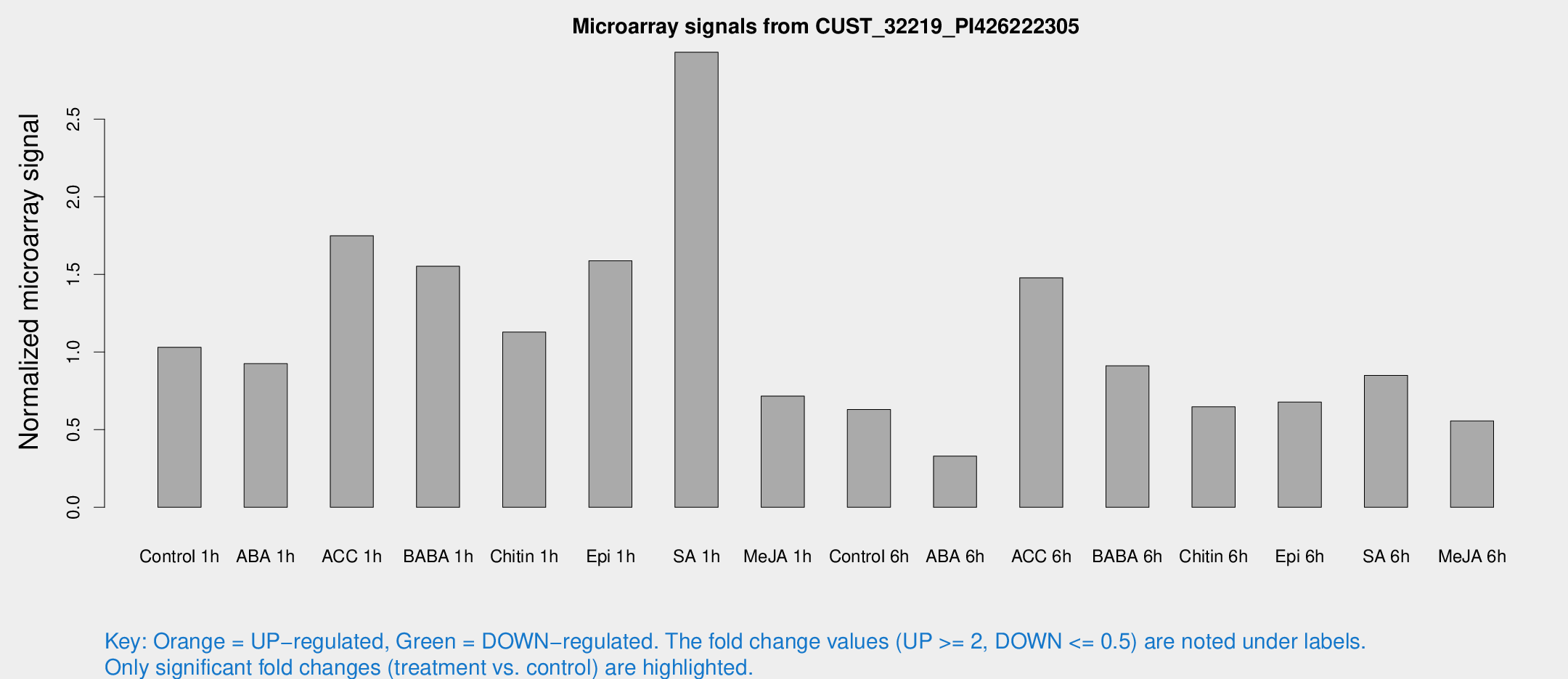
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 136.763 | 22.5943 | 1.03078 | 0.120839 |
| ABA 1h | 106.06 | 7.05965 | 0.925055 | 0.104293 |
| ACC 1h | 294.83 | 109.716 | 1.74883 | 0.98479 |
| BABA 1h | 205.386 | 50.7352 | 1.55255 | 0.267763 |
| Chitin 1h | 134.797 | 22.7976 | 1.12857 | 0.100781 |
| Epi 1h | 178.008 | 10.8976 | 1.58842 | 0.0972068 |
| SA 1h | 408.345 | 81.9763 | 2.93121 | 0.503417 |
| Me-JA 1h | 78.2194 | 13.8777 | 0.716436 | 0.174388 |
| Control 6h | 96.2081 | 31.4649 | 0.630219 | 0.241696 |
| ABA 6h | 59.0924 | 27.497 | 0.329936 | 0.193939 |
| ACC 6h | 220.266 | 14.1272 | 1.4785 | 0.199616 |
| BABA 6h | 145.909 | 44.6288 | 0.91155 | 0.274175 |
| Chitin 6h | 92.2352 | 17.7221 | 0.647506 | 0.103176 |
| Epi 6h | 105.069 | 25.0973 | 0.67791 | 0.146477 |
| SA 6h | 117.161 | 32.1336 | 0.849429 | 0.369889 |
| Me-JA 6h | 75.2192 | 16.2962 | 0.556388 | 0.0774749 |
Source Transcript PGSC0003DMT400012610 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G27290.1 | +3 | 0.0 | 439 | 210/418 (50%) | S-locus lectin protein kinase family protein | chr4:13666281-13669202 FORWARD LENGTH=783 |
