Probe CUST_32050_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32050_PI426222305 | JHI_St_60k_v1 | DMT400013259 | TTGCTGAAGAGCACAAACCAAATTTCAACATCACAGAGATAAAAGCAAATCAGAAGTAGA |
All Microarray Probes Designed to Gene DMG400005179
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32028_PI426222305 | JHI_St_60k_v1 | DMT400013258 | TGCCAGAGGATGAAAAGCACAAACCAAATTTCAACATCACAGAGATAAAAGCAAATCAGA |
| CUST_31980_PI426222305 | JHI_St_60k_v1 | DMT400013257 | CGTCGCAATGATTTGACAGACCAGCTTACCAATTCCAAACAGGTAGTAATTATAGTAGAA |
| CUST_32050_PI426222305 | JHI_St_60k_v1 | DMT400013259 | TTGCTGAAGAGCACAAACCAAATTTCAACATCACAGAGATAAAAGCAAATCAGAAGTAGA |
| CUST_32038_PI426222305 | JHI_St_60k_v1 | DMT400013261 | GGCAGAGTTATCCAGTACCAAAGAATCCTTGTGGTATTTTAATGTTATCTAGATGTTTTC |
| CUST_32044_PI426222305 | JHI_St_60k_v1 | DMT400013260 | GGCTAACTCTAGAAAATTTTCATGTGGCACTGCAATTACCTTACGAAATTTATGCTGTAA |
Microarray Signals from CUST_32050_PI426222305
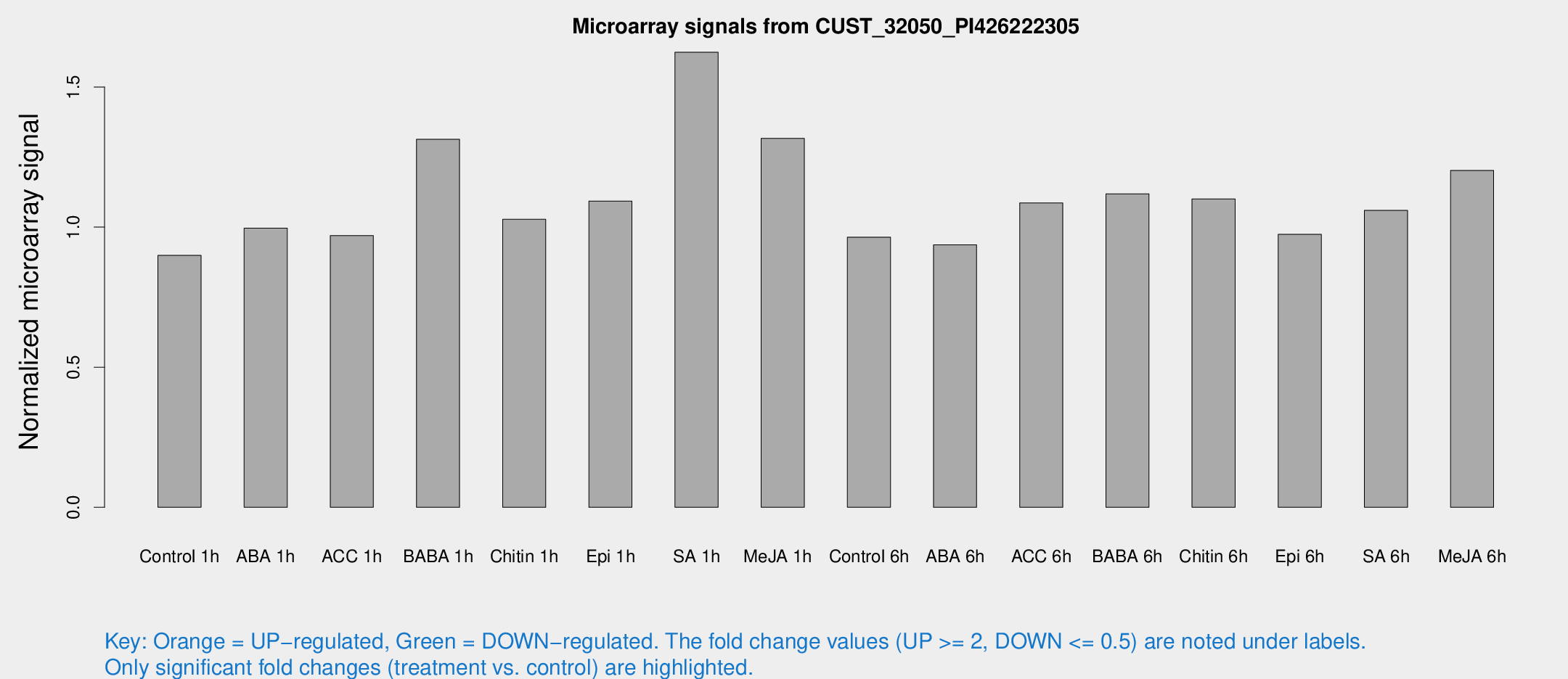
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.97878 | 3.4669 | 0.899023 | 0.521172 |
| ABA 1h | 5.87074 | 3.40645 | 0.996232 | 0.576909 |
| ACC 1h | 6.63995 | 3.85998 | 0.969609 | 0.561894 |
| BABA 1h | 8.69686 | 3.76789 | 1.31336 | 0.610127 |
| Chitin 1h | 6.16226 | 3.58137 | 1.02789 | 0.595744 |
| Epi 1h | 6.32764 | 3.52462 | 1.09276 | 0.611125 |
| SA 1h | 16.0987 | 9.93754 | 1.62409 | 1.47793 |
| Me-JA 1h | 7.41974 | 3.60704 | 1.31689 | 0.676948 |
| Control 6h | 6.42667 | 3.72406 | 0.964233 | 0.558594 |
| ABA 6h | 6.62941 | 3.84553 | 0.936775 | 0.542715 |
| ACC 6h | 8.4316 | 4.56061 | 1.08617 | 0.572879 |
| BABA 6h | 8.71152 | 4.15783 | 1.1188 | 0.569008 |
| Chitin 6h | 7.89729 | 4.03615 | 1.10066 | 0.577898 |
| Epi 6h | 7.36018 | 4.29596 | 0.974095 | 0.564056 |
| SA 6h | 6.99497 | 3.87867 | 1.05944 | 0.590206 |
| Me-JA 6h | 8.45116 | 3.71383 | 1.20235 | 0.590344 |
Source Transcript PGSC0003DMT400013259 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G09900.1 | +2 | 3e-89 | 283 | 177/299 (59%) | methyl esterase 12 | chr4:6221767-6223924 REVERSE LENGTH=349 |
