Probe CUST_31525_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31525_PI426222305 | JHI_St_60k_v1 | DMT400073465 | TATCAATTGCAGAGCTTGAACTAACCAACAATAATCAAAAATCTCCACGCCAAAACCCTC |
All Microarray Probes Designed to Gene DMG400028538
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31474_PI426222305 | JHI_St_60k_v1 | DMT400073464 | TATCAATTGCAGAGCTTGAACTAACCAACAATAATCAAAAATCTCCACGCCAAAACCCTC |
| CUST_31525_PI426222305 | JHI_St_60k_v1 | DMT400073465 | TATCAATTGCAGAGCTTGAACTAACCAACAATAATCAAAAATCTCCACGCCAAAACCCTC |
Microarray Signals from CUST_31525_PI426222305
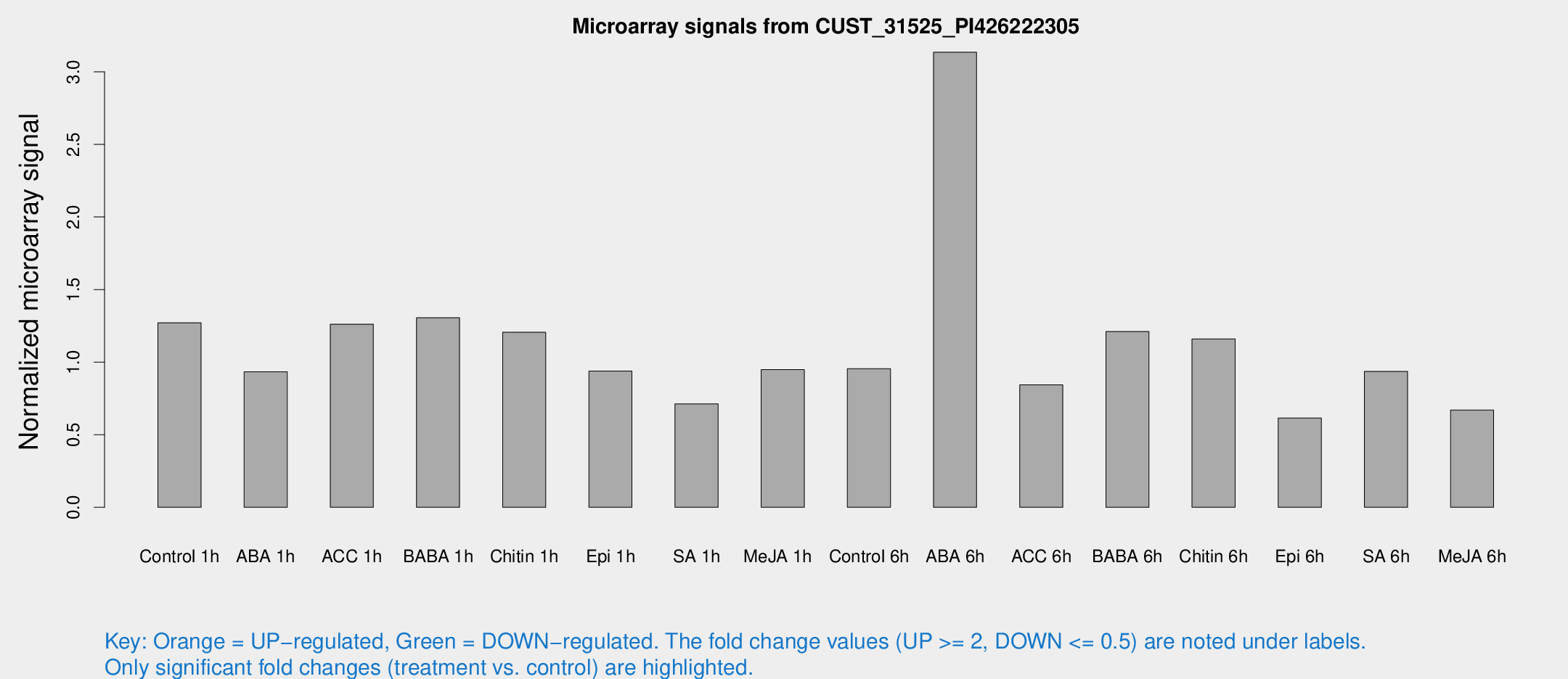
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 13.5665 | 3.52651 | 1.27079 | 0.355238 |
| ABA 1h | 9.06918 | 3.33787 | 0.933951 | 0.391992 |
| ACC 1h | 15.4446 | 5.52423 | 1.26106 | 0.446473 |
| BABA 1h | 14.9321 | 5.39103 | 1.3055 | 0.5697 |
| Chitin 1h | 11.4935 | 3.54553 | 1.20542 | 0.398217 |
| Epi 1h | 8.82963 | 3.40844 | 0.938135 | 0.40125 |
| SA 1h | 8.02296 | 3.38145 | 0.711946 | 0.331014 |
| Me-JA 1h | 8.59721 | 3.58221 | 0.948326 | 0.455396 |
| Control 6h | 10.3682 | 3.79263 | 0.954234 | 0.382232 |
| ABA 6h | 34.5296 | 4.41656 | 3.13477 | 0.401716 |
| ACC 6h | 11.1698 | 4.43611 | 0.843186 | 0.374919 |
| BABA 6h | 16.5713 | 6.43105 | 1.21059 | 0.517553 |
| Chitin 6h | 12.8137 | 4.28098 | 1.16001 | 0.389245 |
| Epi 6h | 7.26495 | 4.27454 | 0.61519 | 0.356856 |
| SA 6h | 9.8722 | 3.99288 | 0.935917 | 0.404522 |
| Me-JA 6h | 6.98626 | 3.76371 | 0.669686 | 0.365413 |
Source Transcript PGSC0003DMT400073465 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G52565.1 | +2 | 1e-10 | 57 | 40/99 (40%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: endomembrane system; BEST Arabidopsis thaliana protein match is: unknown protein (TAIR:AT3G15760.1); Has 35333 Blast hits to 34131 proteins in 2444 species: Archae - 798; Bacteria - 22429; Metazoa - 974; Fungi - 991; Plants - 531; Viruses - 0; Other Eukaryotes - 9610 (source: NCBI BLink). | chr1:19580061-19580511 REVERSE LENGTH=129 |
