Probe CUST_31449_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31449_PI426222305 | JHI_St_60k_v1 | DMT400042530 | TTTGTGAATGTAGGAAAGGACTTTCGGAGTATTATGATGGGAAATCGGAGTCATTTACGT |
All Microarray Probes Designed to Gene DMG400016497
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31536_PI426222305 | JHI_St_60k_v1 | DMT400042531 | CGATTTATCGACTCTTATGGCTCAATTGCCAATCAAGAAAGGACTTTCGGAGTATTATGA |
| CUST_31449_PI426222305 | JHI_St_60k_v1 | DMT400042530 | TTTGTGAATGTAGGAAAGGACTTTCGGAGTATTATGATGGGAAATCGGAGTCATTTACGT |
Microarray Signals from CUST_31449_PI426222305
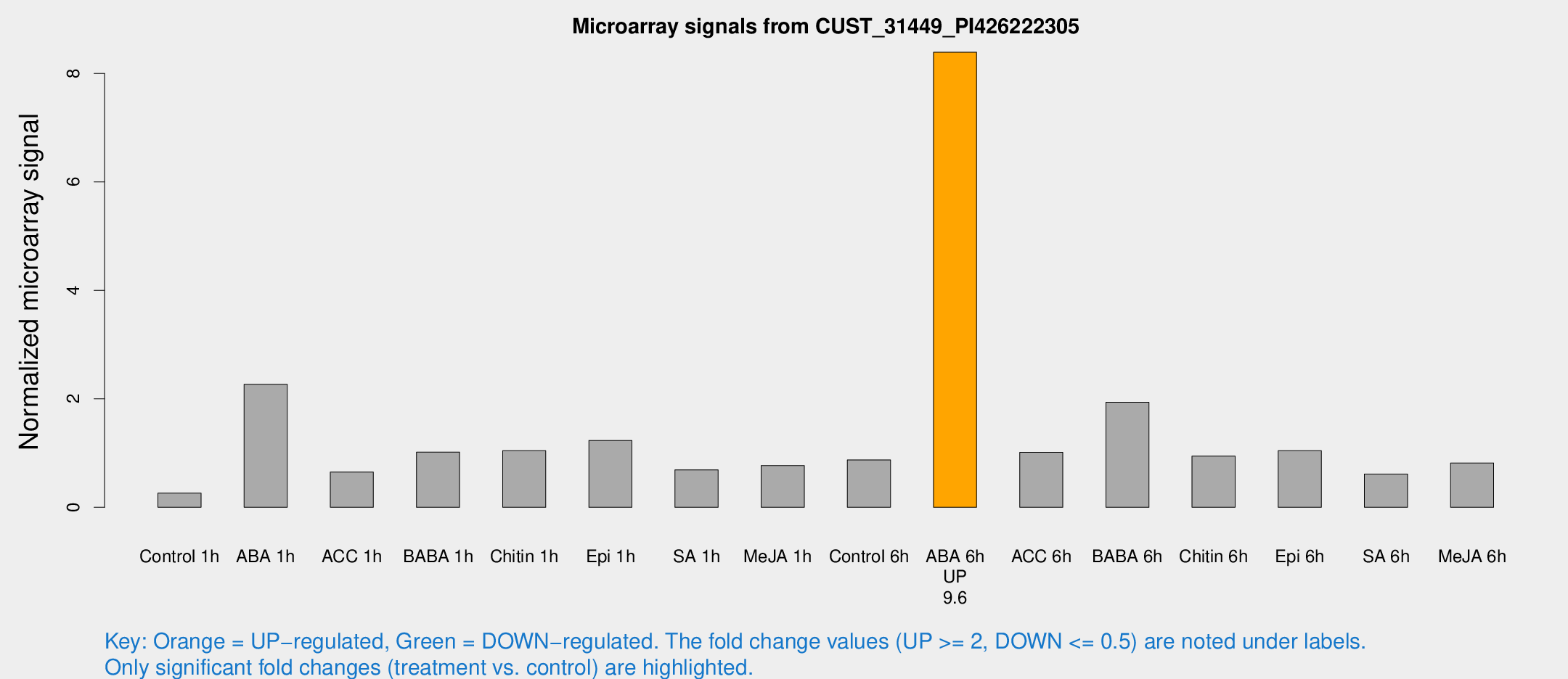
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 11.7239 | 6.05176 | 0.261028 | 0.176793 |
| ABA 1h | 88.4752 | 44.0201 | 2.26705 | 1.02006 |
| ACC 1h | 24.3144 | 4.65903 | 0.65045 | 0.113488 |
| BABA 1h | 36.9523 | 9.14463 | 1.01809 | 0.185178 |
| Chitin 1h | 33.4344 | 4.46252 | 1.04276 | 0.123872 |
| Epi 1h | 43.6194 | 17.2211 | 1.23235 | 0.49682 |
| SA 1h | 25.0797 | 3.5833 | 0.690689 | 0.100419 |
| Me-JA 1h | 23.9095 | 7.0747 | 0.771557 | 0.166371 |
| Control 6h | 38.2533 | 14.1321 | 0.873921 | 0.435454 |
| ABA 6h | 328.573 | 73.015 | 8.39136 | 1.42104 |
| ACC 6h | 46.3656 | 14.7417 | 1.01294 | 0.303241 |
| BABA 6h | 80.0303 | 19.0205 | 1.93703 | 0.378832 |
| Chitin 6h | 37.9197 | 10.0873 | 0.944333 | 0.255114 |
| Epi 6h | 43.7575 | 10.1874 | 1.04435 | 0.218372 |
| SA 6h | 27.8535 | 11.5182 | 0.614412 | 0.333503 |
| Me-JA 6h | 34.2416 | 13.0964 | 0.81758 | 0.363962 |
Source Transcript PGSC0003DMT400042530 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G26288.1 | +2 | 6e-08 | 52 | 30/77 (39%) | FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: cellular_component unknown; BEST Arabidopsis thaliana protein match is: oxidative stress 3 (TAIR:AT5G56550.1); Has 289 Blast hits to 232 proteins in 41 species: Archae - 0; Bacteria - 12; Metazoa - 36; Fungi - 21; Plants - 151; Viruses - 2; Other Eukaryotes - 67 (source: NCBI BLink). | chr4:13307623-13308217 FORWARD LENGTH=155 |
