Probe CUST_3122_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_3122_PI426222305 | JHI_St_60k_v1 | DMT400000446 | CTAGGTGGCTTTATTCAACTTTATTTATTTGGAGAAAGGACTAATCAACCATTGTTGGTC |
All Microarray Probes Designed to Gene DMG400000151
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2920_PI426222305 | JHI_St_60k_v1 | DMT400000443 | GAACTCGGCTTGAAAGTTCCTCCAAAGAGAACAGGCTTTTCTTCTGGAAAATGGATCTAA |
| CUST_2810_PI426222305 | JHI_St_60k_v1 | DMT400000444 | CTAGGTGGCTTTATTCAACTTTATTTATTTGGAGAAAGGACTAATCAACCATTGTTGGTC |
| CUST_3122_PI426222305 | JHI_St_60k_v1 | DMT400000446 | CTAGGTGGCTTTATTCAACTTTATTTATTTGGAGAAAGGACTAATCAACCATTGTTGGTC |
Microarray Signals from CUST_3122_PI426222305
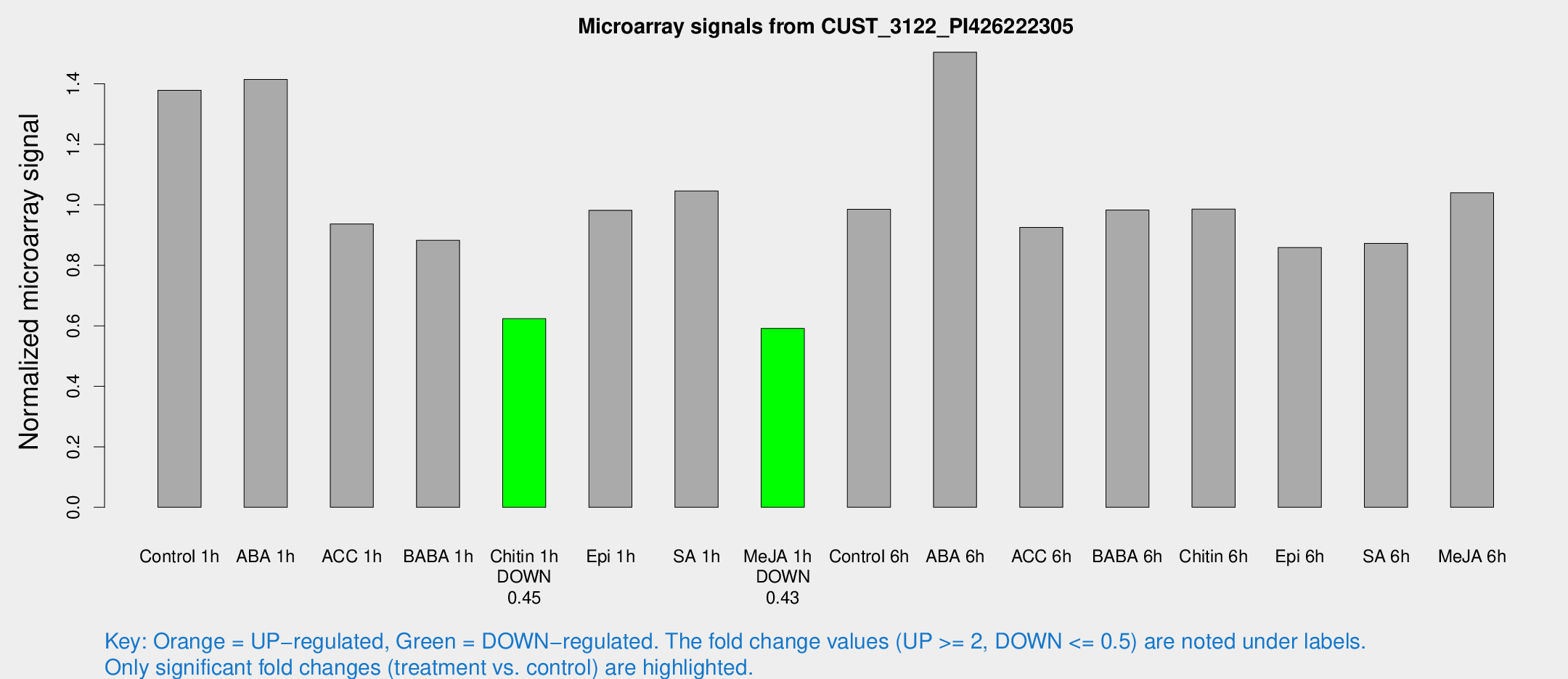
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 684.358 | 87.5953 | 1.3788 | 0.0871376 |
| ABA 1h | 630.239 | 113.231 | 1.41446 | 0.240919 |
| ACC 1h | 561.364 | 195.639 | 0.936764 | 0.406359 |
| BABA 1h | 435.279 | 89.557 | 0.882523 | 0.112317 |
| Chitin 1h | 276.005 | 25.0875 | 0.623624 | 0.0517399 |
| Epi 1h | 426.382 | 66.3733 | 0.981879 | 0.158841 |
| SA 1h | 532.592 | 64.0279 | 1.04597 | 0.0676014 |
| Me-JA 1h | 239.694 | 32.267 | 0.591358 | 0.0406072 |
| Control 6h | 494.872 | 75.5743 | 0.985048 | 0.0779972 |
| ABA 6h | 786.087 | 66.1717 | 1.50446 | 0.0871174 |
| ACC 6h | 522.089 | 43.9117 | 0.925442 | 0.0766014 |
| BABA 6h | 547.549 | 72.7569 | 0.982858 | 0.100691 |
| Chitin 6h | 514.412 | 33.3973 | 0.985859 | 0.0866983 |
| Epi 6h | 476.009 | 39.137 | 0.859031 | 0.0654621 |
| SA 6h | 442.072 | 88.3548 | 0.872252 | 0.097435 |
| Me-JA 6h | 531.399 | 117.398 | 1.03965 | 0.148515 |
Source Transcript PGSC0003DMT400000446 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G00550.1 | +3 | 0.0 | 620 | 300/460 (65%) | digalactosyl diacylglycerol deficient 2 | chr4:238154-240019 REVERSE LENGTH=473 |
