Probe CUST_30455_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30455_PI426222305 | JHI_St_60k_v1 | DMT400069800 | GAGTCCAATGAAACGTTTAGTATAGTAACCATCAAATGCTTGTAAATTCCGACTCTGATT |
All Microarray Probes Designed to Gene DMG400027143
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30292_PI426222305 | JHI_St_60k_v1 | DMT400069798 | TTAAGGATTATGCCTTCTTATTTTAAGATGCGGTACCTTTTGGAAGGTGCATCCTCACCA |
| CUST_30384_PI426222305 | JHI_St_60k_v1 | DMT400069803 | GAGTCCAATGAAACGTTTAGTATAGTAACCATCAAATGCTTGTAAATTCCGACTCTGATT |
| CUST_30455_PI426222305 | JHI_St_60k_v1 | DMT400069800 | GAGTCCAATGAAACGTTTAGTATAGTAACCATCAAATGCTTGTAAATTCCGACTCTGATT |
| CUST_30464_PI426222305 | JHI_St_60k_v1 | DMT400069799 | GAGTCCAATGAAACGTTTAGTATAGTAACCATCAAATGCTTGTAAATTCCGACTCTGATT |
Microarray Signals from CUST_30455_PI426222305
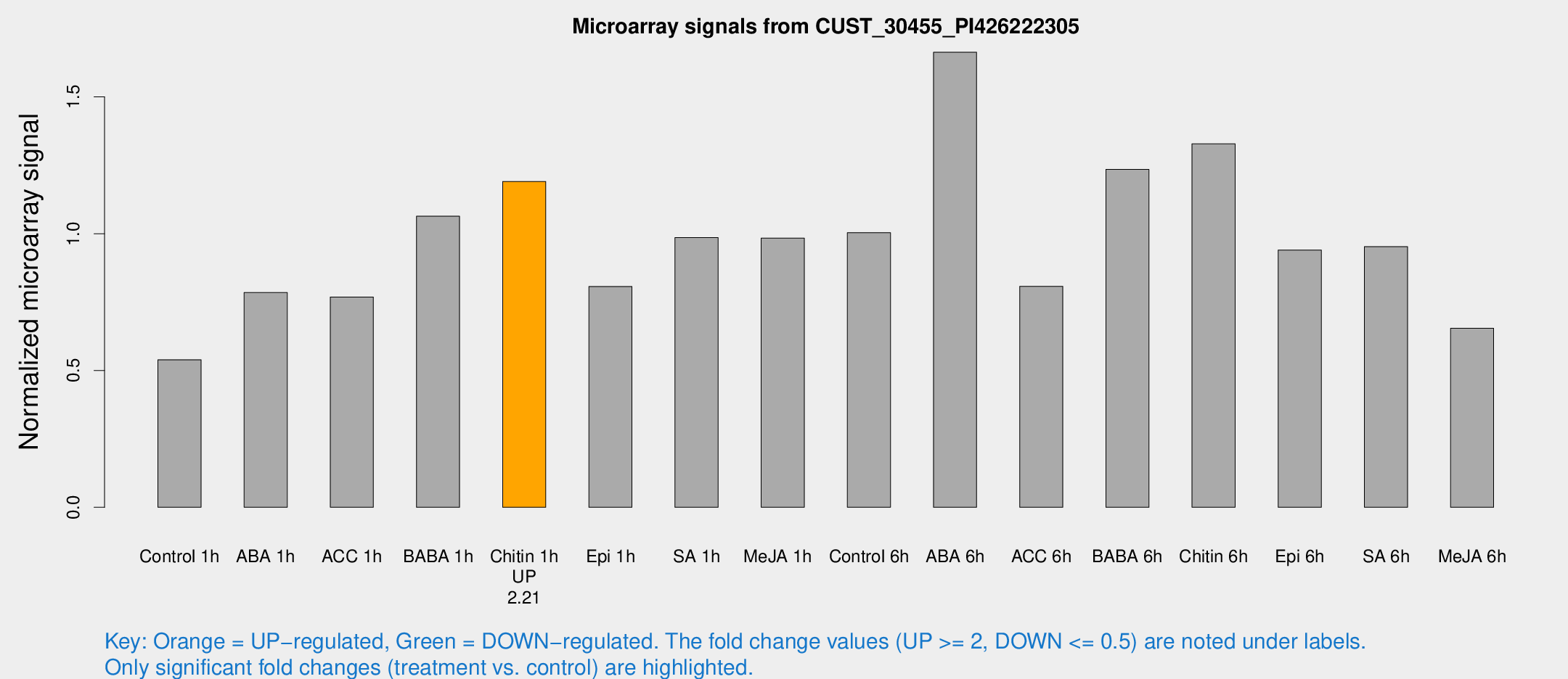
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 304.373 | 28.0436 | 0.539439 | 0.0318032 |
| ABA 1h | 406.181 | 86.7952 | 0.784931 | 0.112378 |
| ACC 1h | 460.669 | 97.9963 | 0.768768 | 0.116217 |
| BABA 1h | 603.578 | 124.084 | 1.06412 | 0.141037 |
| Chitin 1h | 609.755 | 78.7938 | 1.19101 | 0.121694 |
| Epi 1h | 398.168 | 56.4508 | 0.807254 | 0.0883359 |
| SA 1h | 583.99 | 104.222 | 0.985838 | 0.142181 |
| Me-JA 1h | 459.429 | 68.8234 | 0.984211 | 0.118904 |
| Control 6h | 606.124 | 149.061 | 1.00406 | 0.205 |
| ABA 6h | 1000.91 | 109.32 | 1.66339 | 0.148035 |
| ACC 6h | 539.79 | 109.17 | 0.807872 | 0.0649103 |
| BABA 6h | 787.373 | 106.32 | 1.23511 | 0.14198 |
| Chitin 6h | 812.057 | 135.786 | 1.32848 | 0.173539 |
| Epi 6h | 594.537 | 34.7012 | 0.940399 | 0.0547543 |
| SA 6h | 549.523 | 103.924 | 0.952758 | 0.0904654 |
| Me-JA 6h | 415.392 | 125.942 | 0.654302 | 0.219712 |
Source Transcript PGSC0003DMT400069800 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G16950.2 | +3 | 0.0 | 572 | 312/414 (75%) | lipoamide dehydrogenase 1 | chr3:5786508-5790383 REVERSE LENGTH=623 |
