Probe CUST_29221_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29221_PI426222305 | JHI_St_60k_v1 | DMT400053143 | GCGACATAGCTCATAGTTTGAATTTGTTACTTAACCTTTTCTAATCGACTTCACAGGTTA |
All Microarray Probes Designed to Gene DMG400020619
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29186_PI426222305 | JHI_St_60k_v1 | DMT400053142 | ATAAGGTTGGTTTGTATACACTTTATCCTCCCCAGACCCCACTTGACTTGAGGGATTCCA |
| CUST_29221_PI426222305 | JHI_St_60k_v1 | DMT400053143 | GCGACATAGCTCATAGTTTGAATTTGTTACTTAACCTTTTCTAATCGACTTCACAGGTTA |
| CUST_29238_PI426222305 | JHI_St_60k_v1 | DMT400053144 | CAAAAGACACACACGCATCTTATTCTTTGCAAGAACCATCTGAGGTTATGGTGTTTCTAC |
Microarray Signals from CUST_29221_PI426222305
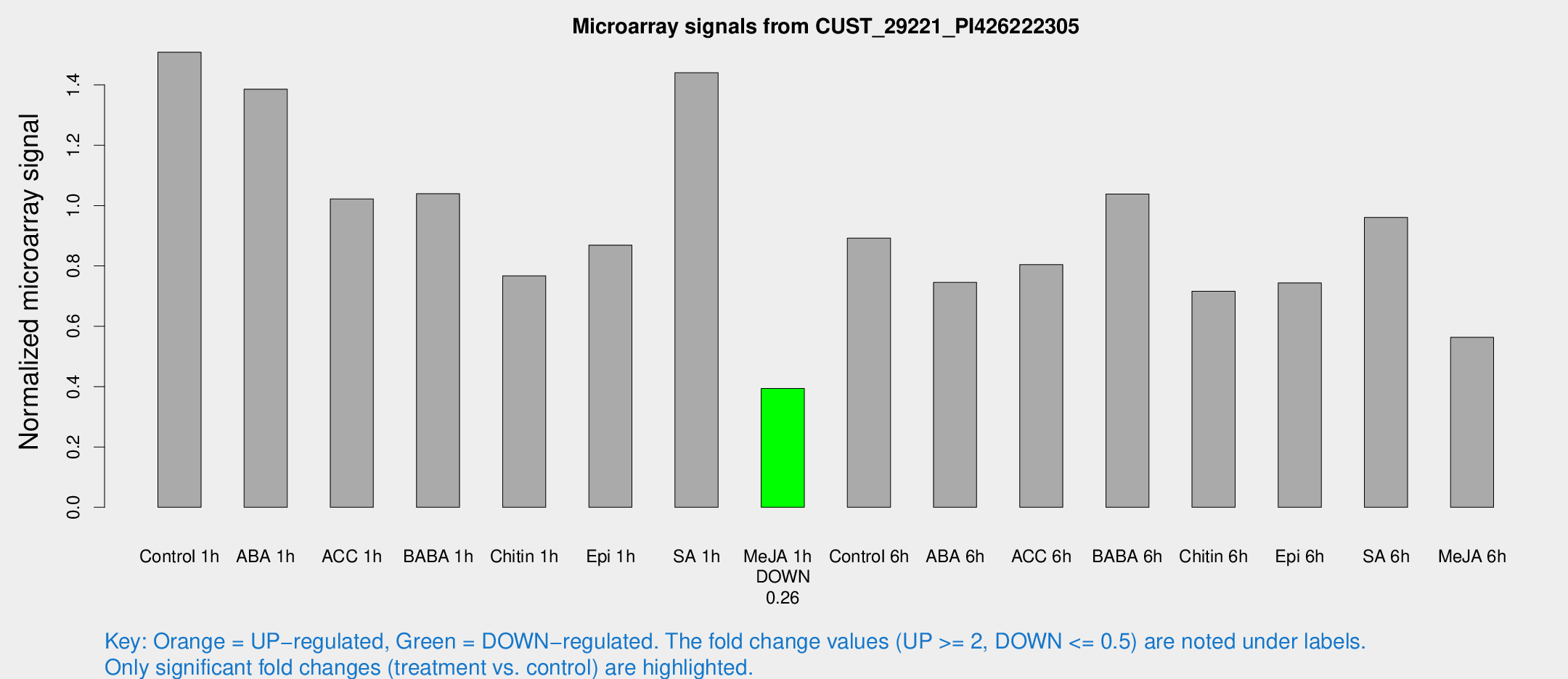
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 46.8871 | 7.50265 | 1.50826 | 0.144818 |
| ABA 1h | 37.2021 | 3.94543 | 1.38587 | 0.147253 |
| ACC 1h | 34.1657 | 8.2356 | 1.02228 | 0.218122 |
| BABA 1h | 36.7456 | 12.7707 | 1.0393 | 0.441272 |
| Chitin 1h | 21.8995 | 4.95024 | 0.767354 | 0.139996 |
| Epi 1h | 23.0962 | 3.62743 | 0.868796 | 0.142666 |
| SA 1h | 49.8231 | 13.8535 | 1.44056 | 0.451141 |
| Me-JA 1h | 9.81991 | 3.58858 | 0.393658 | 0.14453 |
| Control 6h | 29.1113 | 7.08334 | 0.892235 | 0.169407 |
| ABA 6h | 33.0338 | 13.8207 | 0.7455 | 0.599966 |
| ACC 6h | 28.3874 | 4.6634 | 0.804327 | 0.132692 |
| BABA 6h | 38.4605 | 10.455 | 1.03853 | 0.296 |
| Chitin 6h | 25.6365 | 8.59914 | 0.716063 | 0.265074 |
| Epi 6h | 27.849 | 8.75224 | 0.743628 | 0.254698 |
| SA 6h | 28.9736 | 4.18548 | 0.960798 | 0.141189 |
| Me-JA 6h | 21.5051 | 8.66158 | 0.563203 | 0.295433 |
Source Transcript PGSC0003DMT400053143 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G65140.1 | +3 | 2e-156 | 456 | 249/359 (69%) | Haloacid dehalogenase-like hydrolase (HAD) superfamily protein | chr5:26019878-26022077 REVERSE LENGTH=370 |
