Probe CUST_2920_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2920_PI426222305 | JHI_St_60k_v1 | DMT400000443 | GAACTCGGCTTGAAAGTTCCTCCAAAGAGAACAGGCTTTTCTTCTGGAAAATGGATCTAA |
All Microarray Probes Designed to Gene DMG400000151
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2920_PI426222305 | JHI_St_60k_v1 | DMT400000443 | GAACTCGGCTTGAAAGTTCCTCCAAAGAGAACAGGCTTTTCTTCTGGAAAATGGATCTAA |
| CUST_2810_PI426222305 | JHI_St_60k_v1 | DMT400000444 | CTAGGTGGCTTTATTCAACTTTATTTATTTGGAGAAAGGACTAATCAACCATTGTTGGTC |
| CUST_3122_PI426222305 | JHI_St_60k_v1 | DMT400000446 | CTAGGTGGCTTTATTCAACTTTATTTATTTGGAGAAAGGACTAATCAACCATTGTTGGTC |
Microarray Signals from CUST_2920_PI426222305
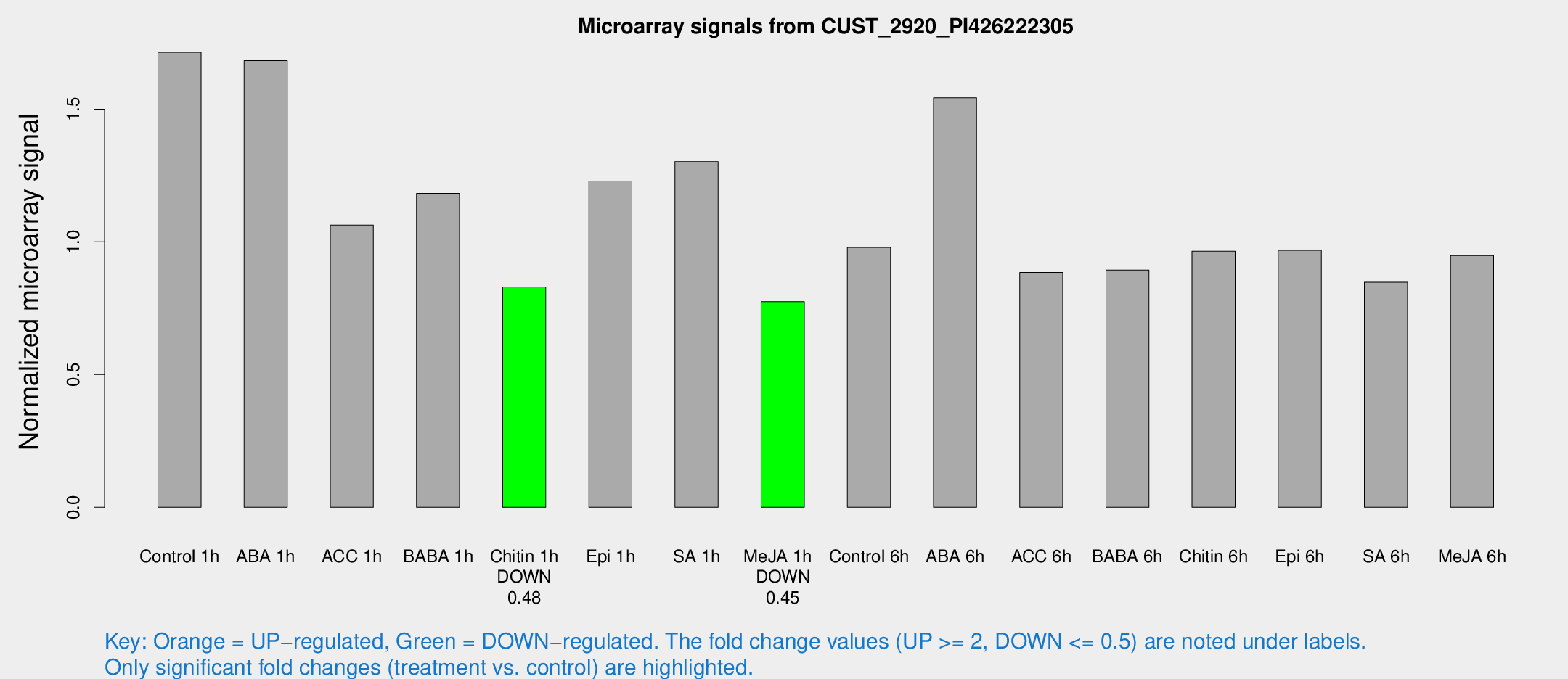
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1882.47 | 169.063 | 1.71418 | 0.0990084 |
| ABA 1h | 1674.1 | 289.144 | 1.68274 | 0.262291 |
| ACC 1h | 1490.16 | 543.419 | 1.06318 | 0.569921 |
| BABA 1h | 1304.05 | 276.612 | 1.18245 | 0.158409 |
| Chitin 1h | 819.63 | 72.9596 | 0.82999 | 0.0587004 |
| Epi 1h | 1192.74 | 190.774 | 1.22921 | 0.204636 |
| SA 1h | 1472.78 | 141.496 | 1.30257 | 0.0760212 |
| Me-JA 1h | 693.137 | 51.4382 | 0.774859 | 0.057816 |
| Control 6h | 1123.33 | 239.402 | 0.97947 | 0.151836 |
| ABA 6h | 1789.73 | 103.534 | 1.54243 | 0.0890957 |
| ACC 6h | 1107.2 | 64.1155 | 0.884634 | 0.0959477 |
| BABA 6h | 1094.72 | 68.0574 | 0.893793 | 0.0516829 |
| Chitin 6h | 1131.11 | 117.857 | 0.964496 | 0.0915833 |
| Epi 6h | 1211.53 | 156.542 | 0.968098 | 0.0838216 |
| SA 6h | 1057.93 | 355.609 | 0.847927 | 0.264688 |
| Me-JA 6h | 1062.03 | 187.387 | 0.948718 | 0.0886214 |
Source Transcript PGSC0003DMT400000443 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G00550.1 | +1 | 0.0 | 534 | 260/423 (61%) | digalactosyl diacylglycerol deficient 2 | chr4:238154-240019 REVERSE LENGTH=473 |
