Probe CUST_2810_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2810_PI426222305 | JHI_St_60k_v1 | DMT400000444 | CTAGGTGGCTTTATTCAACTTTATTTATTTGGAGAAAGGACTAATCAACCATTGTTGGTC |
All Microarray Probes Designed to Gene DMG400000151
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2920_PI426222305 | JHI_St_60k_v1 | DMT400000443 | GAACTCGGCTTGAAAGTTCCTCCAAAGAGAACAGGCTTTTCTTCTGGAAAATGGATCTAA |
| CUST_2810_PI426222305 | JHI_St_60k_v1 | DMT400000444 | CTAGGTGGCTTTATTCAACTTTATTTATTTGGAGAAAGGACTAATCAACCATTGTTGGTC |
| CUST_3122_PI426222305 | JHI_St_60k_v1 | DMT400000446 | CTAGGTGGCTTTATTCAACTTTATTTATTTGGAGAAAGGACTAATCAACCATTGTTGGTC |
Microarray Signals from CUST_2810_PI426222305
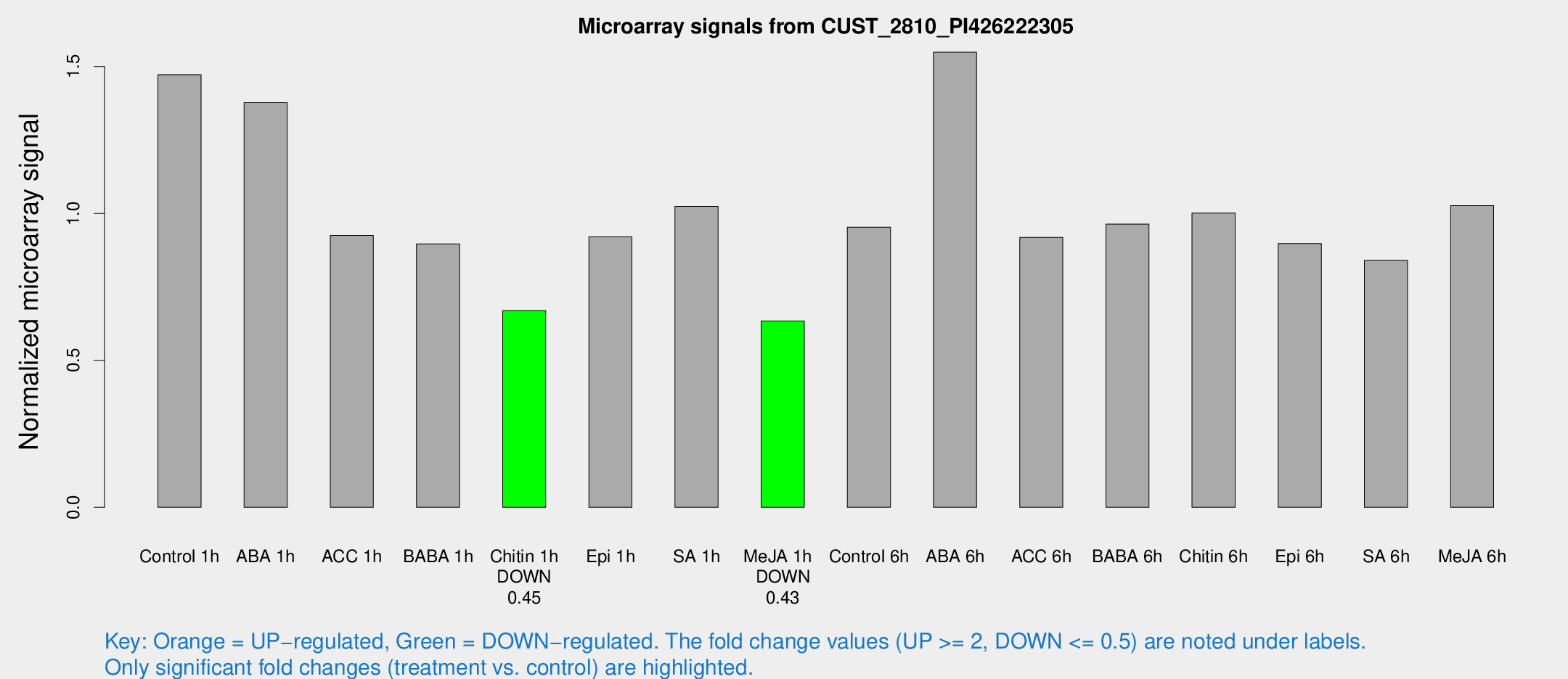
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 697.41 | 102.323 | 1.47235 | 0.127522 |
| ABA 1h | 582.986 | 103.661 | 1.37693 | 0.223468 |
| ACC 1h | 542.805 | 199.126 | 0.925326 | 0.4616 |
| BABA 1h | 420.117 | 86.32 | 0.896389 | 0.115189 |
| Chitin 1h | 279.676 | 16.8065 | 0.669213 | 0.0396079 |
| Epi 1h | 376.135 | 49.473 | 0.920362 | 0.119837 |
| SA 1h | 490.085 | 36.9581 | 1.02395 | 0.0596278 |
| Me-JA 1h | 241.084 | 18.7743 | 0.633786 | 0.0504013 |
| Control 6h | 463.494 | 93.0047 | 0.95336 | 0.133938 |
| ABA 6h | 770.938 | 82.2756 | 1.54872 | 0.112 |
| ACC 6h | 489.388 | 28.6901 | 0.918285 | 0.0992777 |
| BABA 6h | 508.495 | 65.186 | 0.963844 | 0.0908042 |
| Chitin 6h | 494.535 | 28.9223 | 1.00154 | 0.0584942 |
| Epi 6h | 473.81 | 47.9339 | 0.897376 | 0.15702 |
| SA 6h | 412.089 | 96.0719 | 0.839907 | 0.134867 |
| Me-JA 6h | 503.859 | 129.889 | 1.02664 | 0.176054 |
Source Transcript PGSC0003DMT400000444 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G00550.1 | +1 | 4e-168 | 486 | 235/368 (64%) | digalactosyl diacylglycerol deficient 2 | chr4:238154-240019 REVERSE LENGTH=473 |
