Probe CUST_27883_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27883_PI426222305 | JHI_St_60k_v1 | DMT400015950 | TAGATTAGTATCAGTTCAGCTCACCGTTGACCTCAAACCTAACTTACCTCGGGAAAGCGA |
All Microarray Probes Designed to Gene DMG400006242
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27863_PI426222305 | JHI_St_60k_v1 | DMT400015953 | CAACAAGCTTACATGAACTACTAGCTGACTTTCAATTGCTGCTTTTTCCTCTTCTTAATA |
| CUST_27852_PI426222305 | JHI_St_60k_v1 | DMT400015951 | CTCTTTGTAAATGTCCCTTAAGCCTATAGCGACATCGAATTCAATGACAATTAACTAATG |
| CUST_27874_PI426222305 | JHI_St_60k_v1 | DMT400015949 | GAAACATAACACATAGAGATCAGTTTGCCATCTTGGCAACAGATGGGGTATGTATTCCCG |
| CUST_27823_PI426222305 | JHI_St_60k_v1 | DMT400015952 | CAACAAGCTTACATGAACTACTAGCTGACTTTCAATTGCTGCTTTTTCCTCTTCTTAATA |
| CUST_27883_PI426222305 | JHI_St_60k_v1 | DMT400015950 | TAGATTAGTATCAGTTCAGCTCACCGTTGACCTCAAACCTAACTTACCTCGGGAAAGCGA |
Microarray Signals from CUST_27883_PI426222305
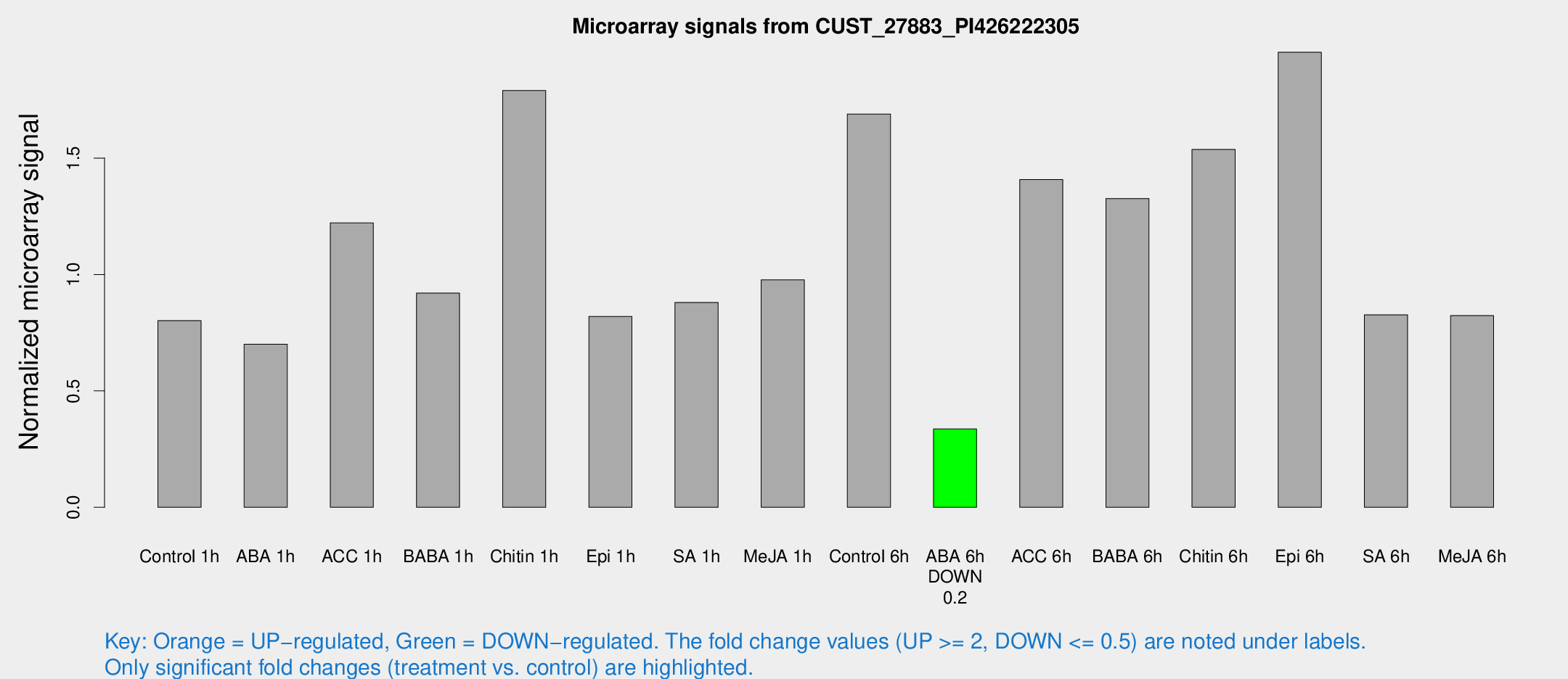
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 115.274 | 21.5092 | 0.801876 | 0.12259 |
| ABA 1h | 87.0744 | 11.4787 | 0.700441 | 0.0866419 |
| ACC 1h | 189.366 | 59.5752 | 1.22141 | 0.500411 |
| BABA 1h | 130.413 | 33.1369 | 0.920013 | 0.164646 |
| Chitin 1h | 260.157 | 101.987 | 1.79015 | 0.742278 |
| Epi 1h | 99.6865 | 13.3345 | 0.819976 | 0.117651 |
| SA 1h | 129.217 | 22.4381 | 0.880056 | 0.122668 |
| Me-JA 1h | 118.177 | 33.0814 | 0.976947 | 0.274157 |
| Control 6h | 281.366 | 98.2323 | 1.68928 | 0.713952 |
| ABA 6h | 49.8867 | 5.1128 | 0.336494 | 0.0329245 |
| ACC 6h | 246.475 | 74.6476 | 1.40781 | 0.304119 |
| BABA 6h | 205.115 | 12.5442 | 1.32636 | 0.0810634 |
| Chitin 6h | 225.99 | 13.6836 | 1.53691 | 0.0929989 |
| Epi 6h | 320.555 | 76.8266 | 1.95479 | 0.444492 |
| SA 6h | 120.357 | 29.2428 | 0.826462 | 0.134052 |
| Me-JA 6h | 121.632 | 29.1197 | 0.823385 | 0.171733 |
Source Transcript PGSC0003DMT400015950 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G05640.1 | +2 | 1e-137 | 422 | 208/355 (59%) | Protein phosphatase 2C family protein | chr3:1640610-1642227 REVERSE LENGTH=358 |
