Probe CUST_27823_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27823_PI426222305 | JHI_St_60k_v1 | DMT400015952 | CAACAAGCTTACATGAACTACTAGCTGACTTTCAATTGCTGCTTTTTCCTCTTCTTAATA |
All Microarray Probes Designed to Gene DMG400006242
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27863_PI426222305 | JHI_St_60k_v1 | DMT400015953 | CAACAAGCTTACATGAACTACTAGCTGACTTTCAATTGCTGCTTTTTCCTCTTCTTAATA |
| CUST_27852_PI426222305 | JHI_St_60k_v1 | DMT400015951 | CTCTTTGTAAATGTCCCTTAAGCCTATAGCGACATCGAATTCAATGACAATTAACTAATG |
| CUST_27874_PI426222305 | JHI_St_60k_v1 | DMT400015949 | GAAACATAACACATAGAGATCAGTTTGCCATCTTGGCAACAGATGGGGTATGTATTCCCG |
| CUST_27823_PI426222305 | JHI_St_60k_v1 | DMT400015952 | CAACAAGCTTACATGAACTACTAGCTGACTTTCAATTGCTGCTTTTTCCTCTTCTTAATA |
| CUST_27883_PI426222305 | JHI_St_60k_v1 | DMT400015950 | TAGATTAGTATCAGTTCAGCTCACCGTTGACCTCAAACCTAACTTACCTCGGGAAAGCGA |
Microarray Signals from CUST_27823_PI426222305
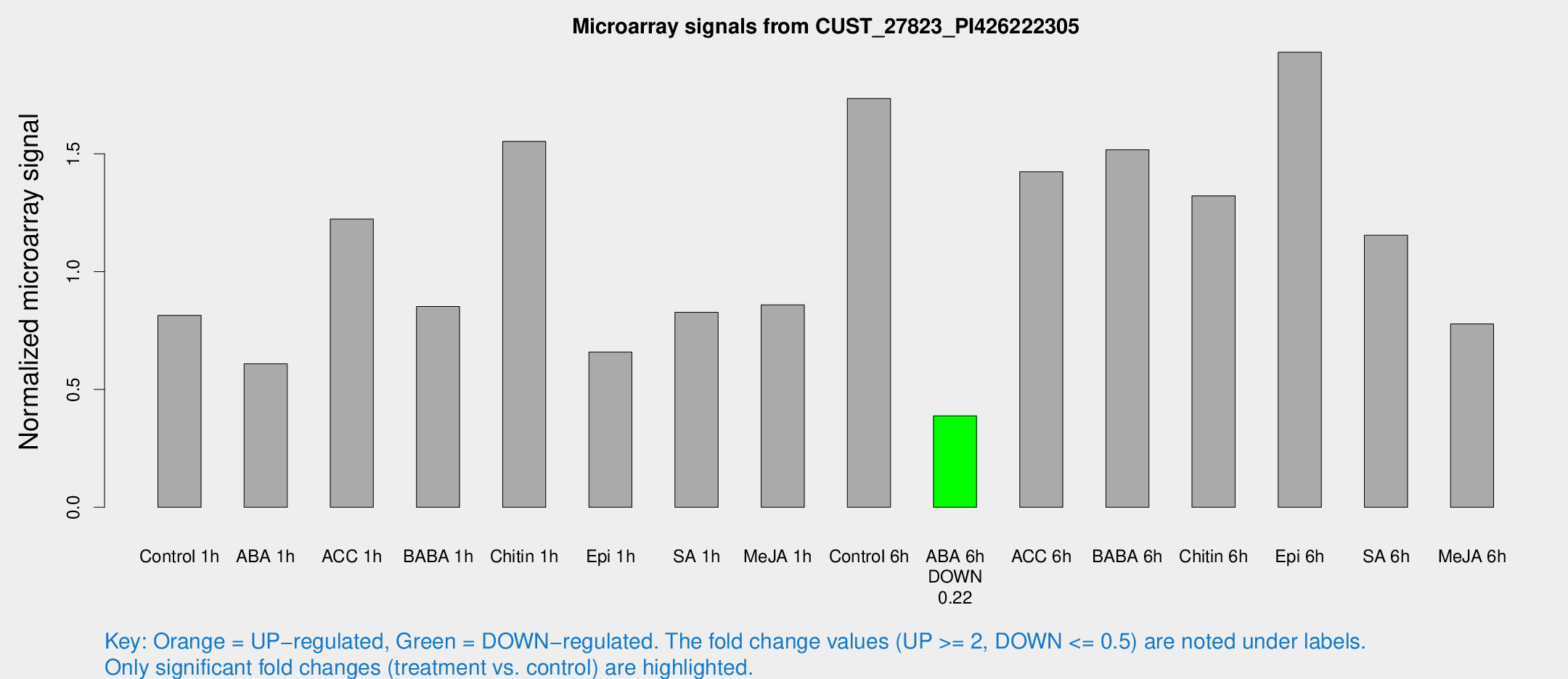
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 263.559 | 19.2122 | 0.814185 | 0.0478806 |
| ABA 1h | 175.802 | 19.7571 | 0.608751 | 0.0900769 |
| ACC 1h | 466.98 | 182.514 | 1.22273 | 0.638275 |
| BABA 1h | 277.856 | 62.0668 | 0.852185 | 0.126679 |
| Chitin 1h | 536.797 | 234.237 | 1.5525 | 0.706894 |
| Epi 1h | 186.893 | 25.8259 | 0.658624 | 0.0984546 |
| SA 1h | 279.62 | 38.7648 | 0.827105 | 0.0848453 |
| Me-JA 1h | 239.215 | 59.9779 | 0.859242 | 0.215768 |
| Control 6h | 627.549 | 190.58 | 1.73446 | 0.476086 |
| ABA 6h | 134.632 | 15.7292 | 0.388197 | 0.0279714 |
| ACC 6h | 552.84 | 117.482 | 1.42387 | 0.187593 |
| BABA 6h | 547.073 | 31.7885 | 1.51657 | 0.0991759 |
| Chitin 6h | 455.152 | 31.4285 | 1.32164 | 0.0974601 |
| Epi 6h | 725.277 | 138.388 | 1.9308 | 0.402207 |
| SA 6h | 376.558 | 54.4113 | 1.15444 | 0.067509 |
| Me-JA 6h | 255.523 | 37.5888 | 0.778004 | 0.0587216 |
Source Transcript PGSC0003DMT400015952 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G05640.1 | +1 | 4e-61 | 185 | 87/130 (67%) | Protein phosphatase 2C family protein | chr3:1640610-1642227 REVERSE LENGTH=358 |
