Probe CUST_26827_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26827_PI426222305 | JHI_St_60k_v1 | DMT400035263 | CCCTTCTGTAGTCATGAAGAGGATTGCAAATTTGACATTATGTAGTGTTACTGTGAATAA |
All Microarray Probes Designed to Gene DMG400013546
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26822_PI426222305 | JHI_St_60k_v1 | DMT400035262 | CCCTTCTGTAGTCATGAAGAGGATTGCAAATTTGACATTATGTAGTGTTACTGTGAATAA |
| CUST_26827_PI426222305 | JHI_St_60k_v1 | DMT400035263 | CCCTTCTGTAGTCATGAAGAGGATTGCAAATTTGACATTATGTAGTGTTACTGTGAATAA |
| CUST_26806_PI426222305 | JHI_St_60k_v1 | DMT400035260 | CTTGAGATGTTTTATGCTCTCAAATTCCGCAAGCTGGCTCAACTTGTTCCATTGGCTGTT |
Microarray Signals from CUST_26827_PI426222305
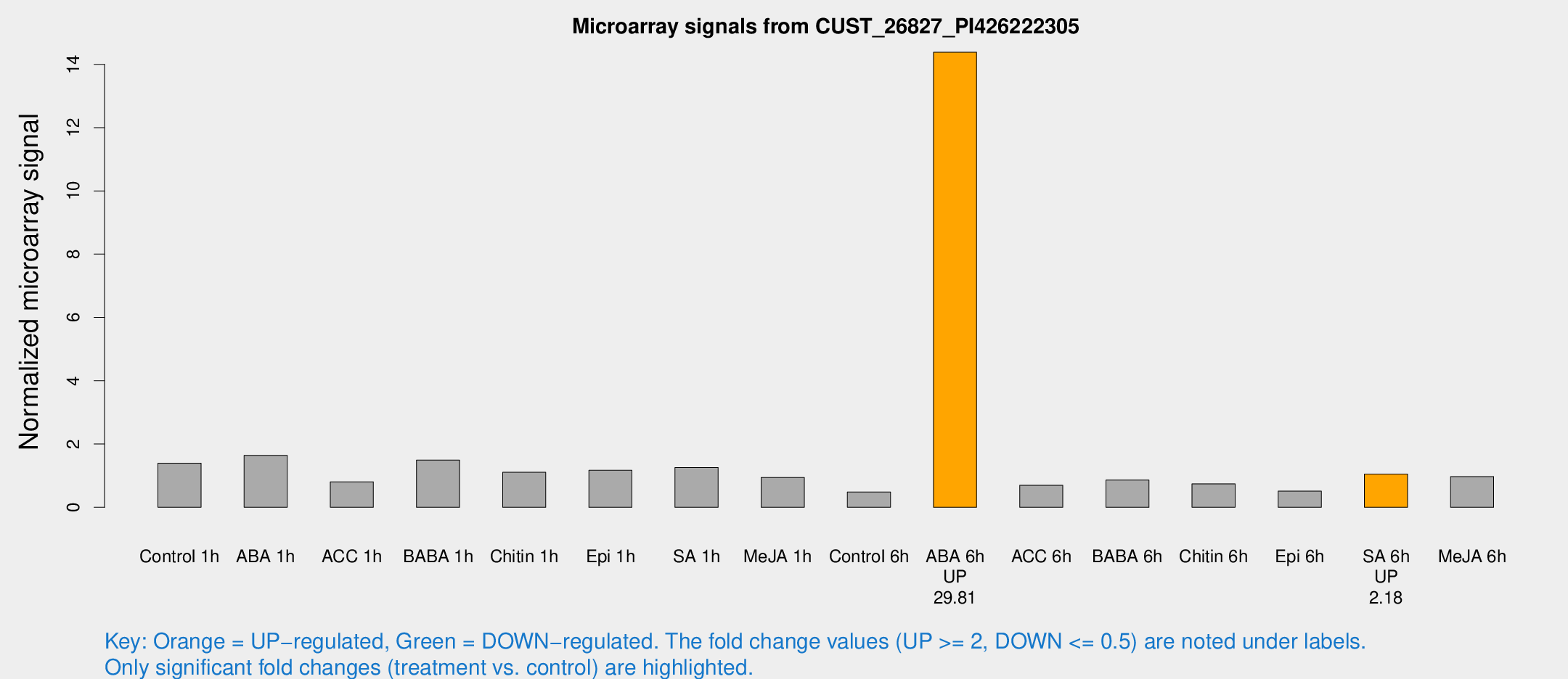
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 884.405 | 114.175 | 1.39311 | 0.134153 |
| ABA 1h | 957.133 | 199.024 | 1.64239 | 0.311915 |
| ACC 1h | 565.087 | 147.376 | 0.805297 | 0.20324 |
| BABA 1h | 918.902 | 132.719 | 1.49178 | 0.0893478 |
| Chitin 1h | 638.197 | 107.518 | 1.10632 | 0.0979993 |
| Epi 1h | 649.707 | 99.3278 | 1.16986 | 0.182484 |
| SA 1h | 829.041 | 124.368 | 1.25945 | 0.137041 |
| Me-JA 1h | 481.909 | 28.0465 | 0.942929 | 0.0547659 |
| Control 6h | 324.622 | 77.7353 | 0.482427 | 0.0954425 |
| ABA 6h | 10158 | 2327.37 | 14.3817 | 3.05143 |
| ACC 6h | 499.671 | 29.1227 | 0.696345 | 0.0766554 |
| BABA 6h | 605.462 | 54.7286 | 0.858923 | 0.049832 |
| Chitin 6h | 494.888 | 30.0148 | 0.74139 | 0.0566639 |
| Epi 6h | 360.736 | 21.3645 | 0.510342 | 0.0704061 |
| SA 6h | 654.709 | 57.758 | 1.05167 | 0.101684 |
| Me-JA 6h | 618.376 | 99.8921 | 0.969459 | 0.0775263 |
Source Transcript PGSC0003DMT400035263 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G43190.1 | +3 | 0.0 | 1219 | 595/721 (83%) | sucrose synthase 4 | chr3:15179204-15182577 REVERSE LENGTH=808 |
