Probe CUST_26822_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26822_PI426222305 | JHI_St_60k_v1 | DMT400035262 | CCCTTCTGTAGTCATGAAGAGGATTGCAAATTTGACATTATGTAGTGTTACTGTGAATAA |
All Microarray Probes Designed to Gene DMG400013546
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26822_PI426222305 | JHI_St_60k_v1 | DMT400035262 | CCCTTCTGTAGTCATGAAGAGGATTGCAAATTTGACATTATGTAGTGTTACTGTGAATAA |
| CUST_26827_PI426222305 | JHI_St_60k_v1 | DMT400035263 | CCCTTCTGTAGTCATGAAGAGGATTGCAAATTTGACATTATGTAGTGTTACTGTGAATAA |
| CUST_26806_PI426222305 | JHI_St_60k_v1 | DMT400035260 | CTTGAGATGTTTTATGCTCTCAAATTCCGCAAGCTGGCTCAACTTGTTCCATTGGCTGTT |
Microarray Signals from CUST_26822_PI426222305
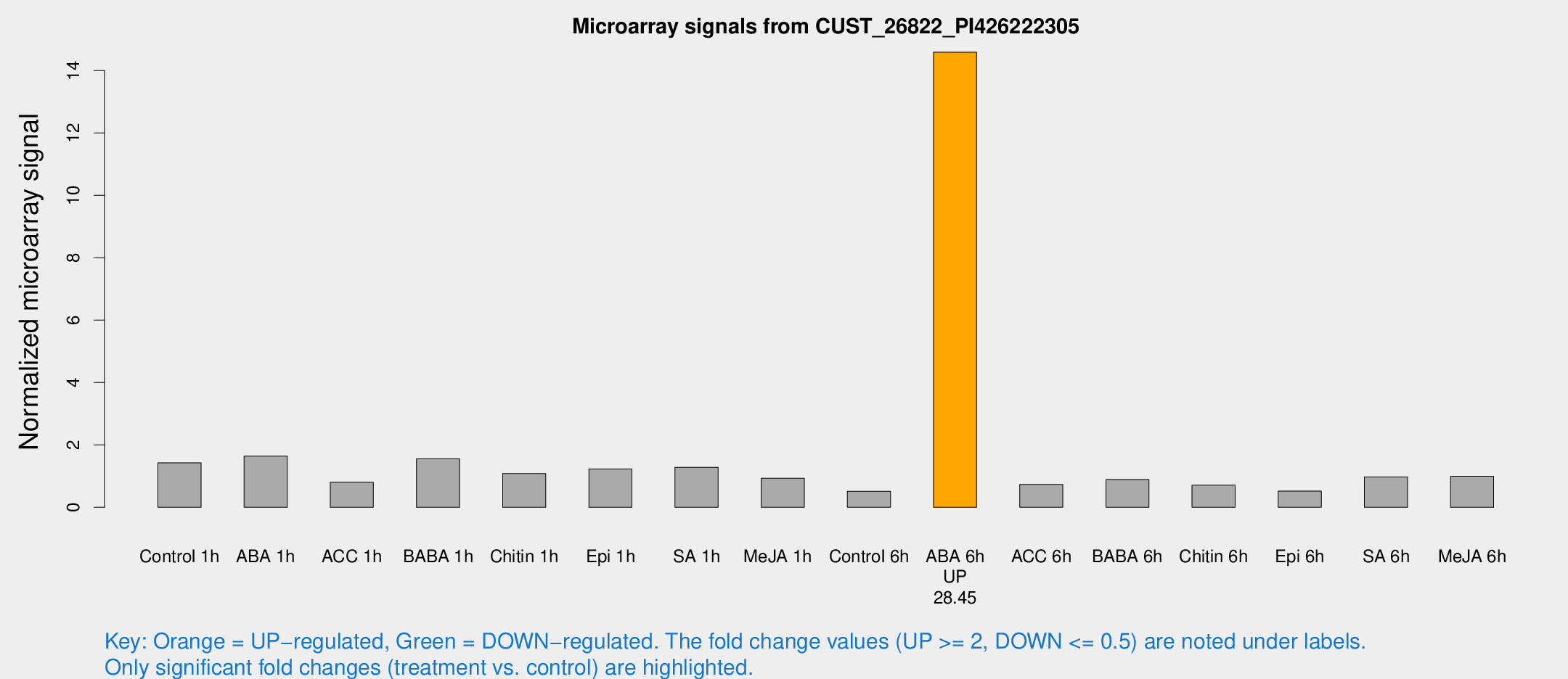
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 885.824 | 134.871 | 1.42368 | 0.125685 |
| ABA 1h | 920.968 | 177.204 | 1.63891 | 0.289097 |
| ACC 1h | 561.205 | 158.667 | 0.804567 | 0.236662 |
| BABA 1h | 933.746 | 147.126 | 1.55164 | 0.138955 |
| Chitin 1h | 613.957 | 120.932 | 1.0815 | 0.1495 |
| Epi 1h | 673.046 | 124.655 | 1.22575 | 0.245708 |
| SA 1h | 828.124 | 139.827 | 1.28355 | 0.152835 |
| Me-JA 1h | 465.114 | 27.2843 | 0.932327 | 0.0542092 |
| Control 6h | 329.049 | 66.6565 | 0.512752 | 0.0725426 |
| ABA 6h | 10022.2 | 2272.4 | 14.5875 | 3.00122 |
| ACC 6h | 512.573 | 29.9431 | 0.731651 | 0.0683144 |
| BABA 6h | 612.287 | 57.4085 | 0.890699 | 0.0535071 |
| Chitin 6h | 460.818 | 26.8435 | 0.710817 | 0.0414022 |
| Epi 6h | 357.41 | 20.985 | 0.520164 | 0.0305133 |
| SA 6h | 595.74 | 71.7947 | 0.975178 | 0.0826106 |
| Me-JA 6h | 623.56 | 118.315 | 0.994797 | 0.127065 |
Source Transcript PGSC0003DMT400035262 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G43190.1 | +1 | 0.0 | 1303 | 640/797 (80%) | sucrose synthase 4 | chr3:15179204-15182577 REVERSE LENGTH=808 |
