Probe CUST_26477_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26477_PI426222305 | JHI_St_60k_v1 | DMT400036997 | CTGAAAATTGACGTTGAACCTGAGCTCCTTAGGGGATATGTCAACCAAGAAACTGGCAAG |
All Microarray Probes Designed to Gene DMG400014262
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26477_PI426222305 | JHI_St_60k_v1 | DMT400036997 | CTGAAAATTGACGTTGAACCTGAGCTCCTTAGGGGATATGTCAACCAAGAAACTGGCAAG |
Microarray Signals from CUST_26477_PI426222305
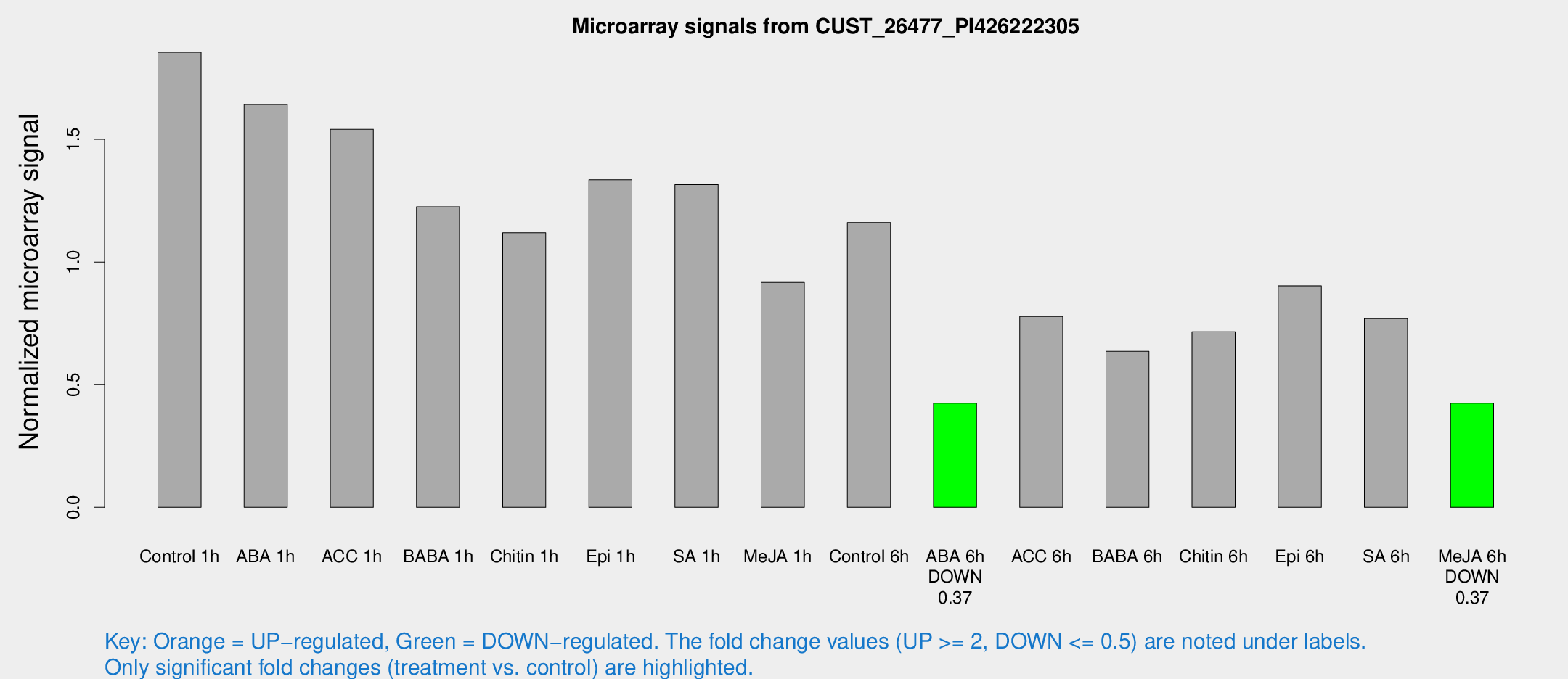
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5250.55 | 859.334 | 1.8549 | 0.229515 |
| ABA 1h | 4023.88 | 255.994 | 1.64224 | 0.215689 |
| ACC 1h | 4429.92 | 525.535 | 1.54052 | 0.0978223 |
| BABA 1h | 3403.07 | 655.614 | 1.2249 | 0.139102 |
| Chitin 1h | 2779.91 | 160.703 | 1.11937 | 0.0646442 |
| Epi 1h | 3217.94 | 304.167 | 1.3351 | 0.123431 |
| SA 1h | 3740.63 | 216.678 | 1.3156 | 0.075967 |
| Me-JA 1h | 2076.63 | 154.805 | 0.916555 | 0.0529425 |
| Control 6h | 3381.77 | 697.691 | 1.16104 | 0.17238 |
| ABA 6h | 1260.18 | 132.323 | 0.424486 | 0.0319143 |
| ACC 6h | 2537.88 | 447.707 | 0.777619 | 0.044919 |
| BABA 6h | 2007.59 | 293.715 | 0.635905 | 0.109062 |
| Chitin 6h | 2147.98 | 328.534 | 0.715618 | 0.0838002 |
| Epi 6h | 2849.68 | 348.475 | 0.902987 | 0.138592 |
| SA 6h | 2168.77 | 365.477 | 0.76879 | 0.053402 |
| Me-JA 6h | 1188.73 | 159.952 | 0.424332 | 0.0245398 |
Source Transcript PGSC0003DMT400036997 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G62140.1 | +3 | 3e-51 | 179 | 123/227 (54%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: chloroplast; EXPRESSED IN: 19 plant structures; EXPRESSED DURING: 13 growth stages; Has 60 Blast hits to 60 proteins in 24 species: Archae - 0; Bacteria - 14; Metazoa - 0; Fungi - 0; Plants - 45; Viruses - 0; Other Eukaryotes - 1 (source: NCBI BLink). | chr5:24954563-24955376 REVERSE LENGTH=241 |
