Probe CUST_23915_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23915_PI426222305 | JHI_St_60k_v1 | DMT400032758 | ACTGCAGGTATGCATGTGATGAATGGGGTGTTGGTATGGAAATTGACAAGAATGTGAAGA |
All Microarray Probes Designed to Gene DMG400012578
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23915_PI426222305 | JHI_St_60k_v1 | DMT400032758 | ACTGCAGGTATGCATGTGATGAATGGGGTGTTGGTATGGAAATTGACAAGAATGTGAAGA |
| CUST_23948_PI426222305 | JHI_St_60k_v1 | DMT400032759 | ATGGAATTCAGTGATGGAGAGTATAGGAAGTGGCGTACCCATGATTTGCTGGCCTTTTTT |
| CUST_23855_PI426222305 | JHI_St_60k_v1 | DMT400032757 | TTTCATGTGCAGGTATGCATGTGATGAATGGGGTGTTGGTATGGAAATTGACAAGAATGT |
Microarray Signals from CUST_23915_PI426222305
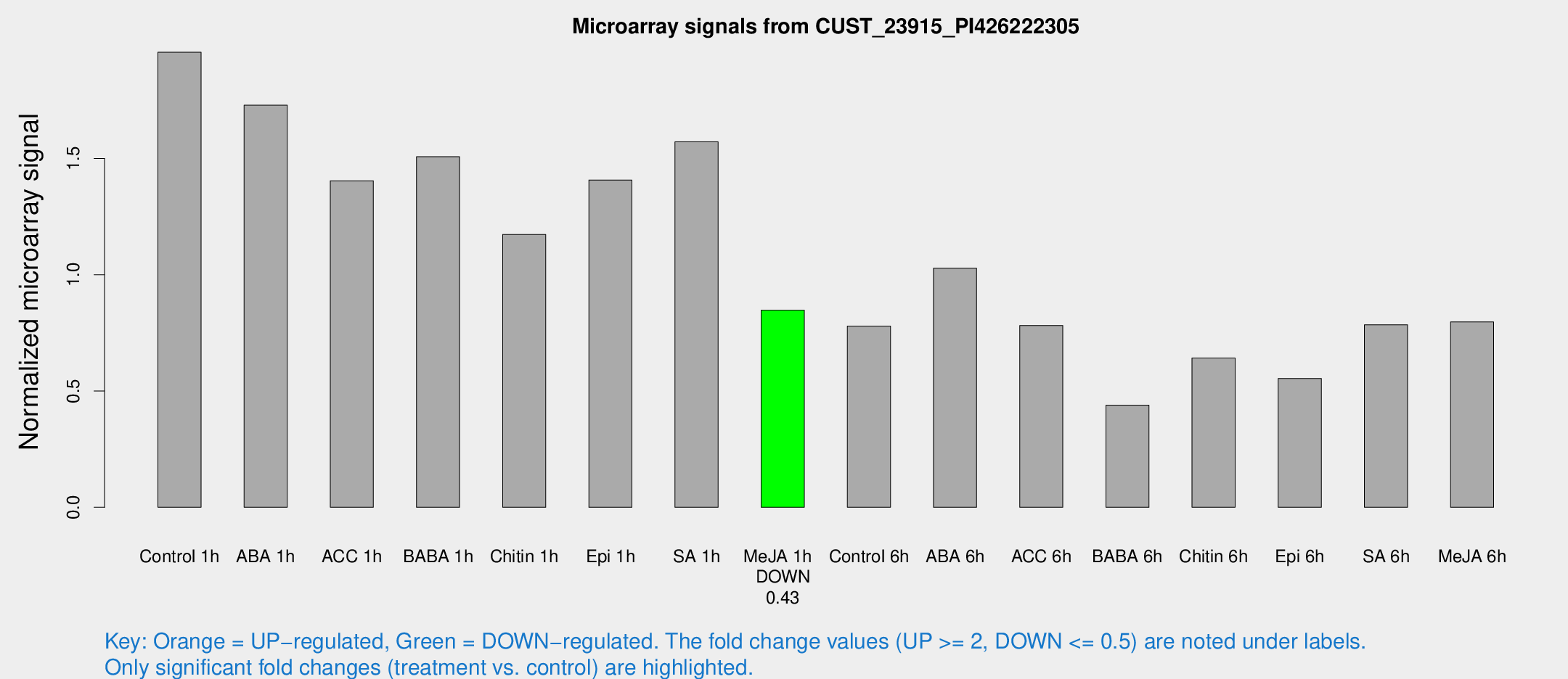
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5844.97 | 405.218 | 1.95713 | 0.207074 |
| ABA 1h | 4634.34 | 645.25 | 1.72902 | 0.331202 |
| ACC 1h | 4668.89 | 1334.47 | 1.40452 | 0.3462 |
| BABA 1h | 4508.83 | 854.491 | 1.50782 | 0.166878 |
| Chitin 1h | 3203.47 | 475.104 | 1.17338 | 0.137888 |
| Epi 1h | 3753.12 | 744.007 | 1.40724 | 0.279763 |
| SA 1h | 5082.73 | 1116.06 | 1.57198 | 0.334723 |
| Me-JA 1h | 2076.48 | 195.796 | 0.848111 | 0.0489825 |
| Control 6h | 2482.92 | 665.138 | 0.779354 | 0.169818 |
| ABA 6h | 3535.93 | 897.501 | 1.02846 | 0.290447 |
| ACC 6h | 2672.6 | 154.74 | 0.782075 | 0.132084 |
| BABA 6h | 1524.38 | 319.225 | 0.43931 | 0.0970157 |
| Chitin 6h | 2042.95 | 147.315 | 0.642315 | 0.0499304 |
| Epi 6h | 1869.9 | 150.816 | 0.554289 | 0.0320198 |
| SA 6h | 2528.62 | 676.265 | 0.785125 | 0.163744 |
| Me-JA 6h | 2475.16 | 515.582 | 0.796986 | 0.185001 |
Source Transcript PGSC0003DMT400032758 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G22340.1 | +3 | 0.0 | 554 | 259/457 (57%) | UDP-glucosyl transferase 85A7 | chr1:7890464-7892090 REVERSE LENGTH=487 |
