Probe CUST_22314_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22314_PI426222305 | JHI_St_60k_v1 | DMT400023397 | TTCGTTGTTTTAATCTCAGCTGAAATTGTTGATAGTCCTCATTTGGATCATCTTCTCATG |
All Microarray Probes Designed to Gene DMG400009061
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22209_PI426222305 | JHI_St_60k_v1 | DMT400023396 | GGAAAGTGCAGCTGGTTTTTCCGACGAGAGCAGTGCAATAAAAGATGACTTATTCGTTGG |
| CUST_22294_PI426222305 | JHI_St_60k_v1 | DMT400023394 | TTTGTTGTTCTTCTGTCAGCTGAGATTATTGATAGCCCTTATTTGTCTTCCATAATCAAG |
| CUST_22314_PI426222305 | JHI_St_60k_v1 | DMT400023397 | TTCGTTGTTTTAATCTCAGCTGAAATTGTTGATAGTCCTCATTTGGATCATCTTCTCATG |
| CUST_22214_PI426222305 | JHI_St_60k_v1 | DMT400023398 | TTGGATATTCCAGCTCTGGAGAAGTTTTGAACTGCAAGAAGTGAGCCCAAGTATGTAGGC |
| CUST_22181_PI426222305 | JHI_St_60k_v1 | DMT400023399 | TTCGTTGTTTTAATCTCAGCTGAAATTGTTGATAGTCCTCATTTGGATCATCTTCTCATG |
Microarray Signals from CUST_22314_PI426222305
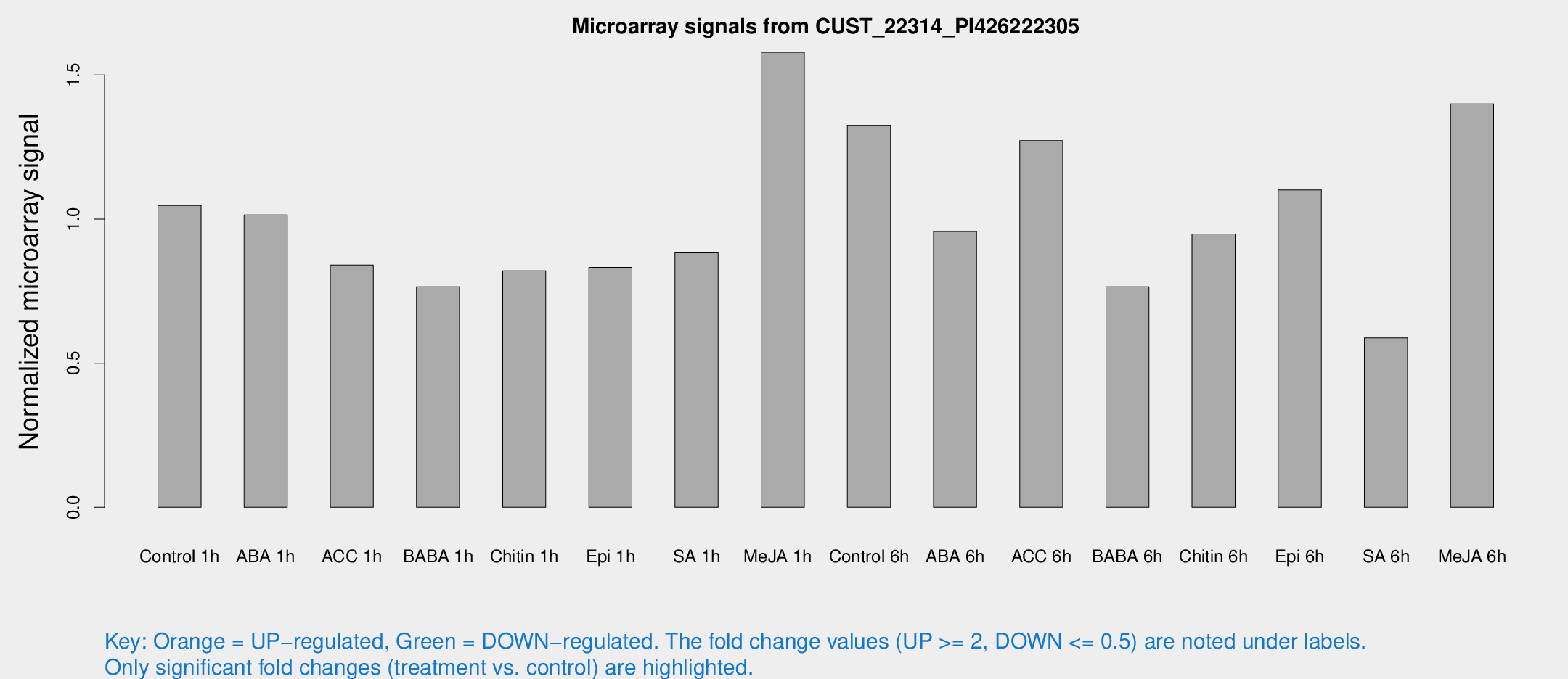
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 49.3201 | 4.19292 | 1.04705 | 0.0875042 |
| ABA 1h | 42.1781 | 3.74983 | 1.01459 | 0.090868 |
| ACC 1h | 40.8431 | 4.26399 | 0.840752 | 0.0882124 |
| BABA 1h | 40.3903 | 12.9915 | 0.765156 | 0.274267 |
| Chitin 1h | 36.2017 | 8.21333 | 0.821003 | 0.184629 |
| Epi 1h | 33.7521 | 3.50533 | 0.832745 | 0.0868011 |
| SA 1h | 42.8887 | 4.59554 | 0.882824 | 0.113652 |
| Me-JA 1h | 60.7923 | 5.29986 | 1.57896 | 0.121036 |
| Control 6h | 66.6939 | 15.9548 | 1.32419 | 0.260982 |
| ABA 6h | 51.5802 | 12.9303 | 0.957471 | 0.262175 |
| ACC 6h | 69.0623 | 7.49988 | 1.27222 | 0.0994051 |
| BABA 6h | 43.3338 | 11.6456 | 0.765471 | 0.195909 |
| Chitin 6h | 48.2306 | 6.63925 | 0.948405 | 0.15049 |
| Epi 6h | 63.7409 | 17.0495 | 1.10108 | 0.471433 |
| SA 6h | 29.2063 | 7.28549 | 0.587682 | 0.206794 |
| Me-JA 6h | 75.1257 | 24.1906 | 1.39962 | 0.487736 |
Source Transcript PGSC0003DMT400023397 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G37670.2 | +3 | 1e-120 | 261 | 139/243 (57%) | N-acetyl-l-glutamate synthase 2 | chr4:17696179-17698836 REVERSE LENGTH=613 |
