Probe CUST_22294_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22294_PI426222305 | JHI_St_60k_v1 | DMT400023394 | TTTGTTGTTCTTCTGTCAGCTGAGATTATTGATAGCCCTTATTTGTCTTCCATAATCAAG |
All Microarray Probes Designed to Gene DMG400009061
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22209_PI426222305 | JHI_St_60k_v1 | DMT400023396 | GGAAAGTGCAGCTGGTTTTTCCGACGAGAGCAGTGCAATAAAAGATGACTTATTCGTTGG |
| CUST_22294_PI426222305 | JHI_St_60k_v1 | DMT400023394 | TTTGTTGTTCTTCTGTCAGCTGAGATTATTGATAGCCCTTATTTGTCTTCCATAATCAAG |
| CUST_22314_PI426222305 | JHI_St_60k_v1 | DMT400023397 | TTCGTTGTTTTAATCTCAGCTGAAATTGTTGATAGTCCTCATTTGGATCATCTTCTCATG |
| CUST_22214_PI426222305 | JHI_St_60k_v1 | DMT400023398 | TTGGATATTCCAGCTCTGGAGAAGTTTTGAACTGCAAGAAGTGAGCCCAAGTATGTAGGC |
| CUST_22181_PI426222305 | JHI_St_60k_v1 | DMT400023399 | TTCGTTGTTTTAATCTCAGCTGAAATTGTTGATAGTCCTCATTTGGATCATCTTCTCATG |
Microarray Signals from CUST_22294_PI426222305
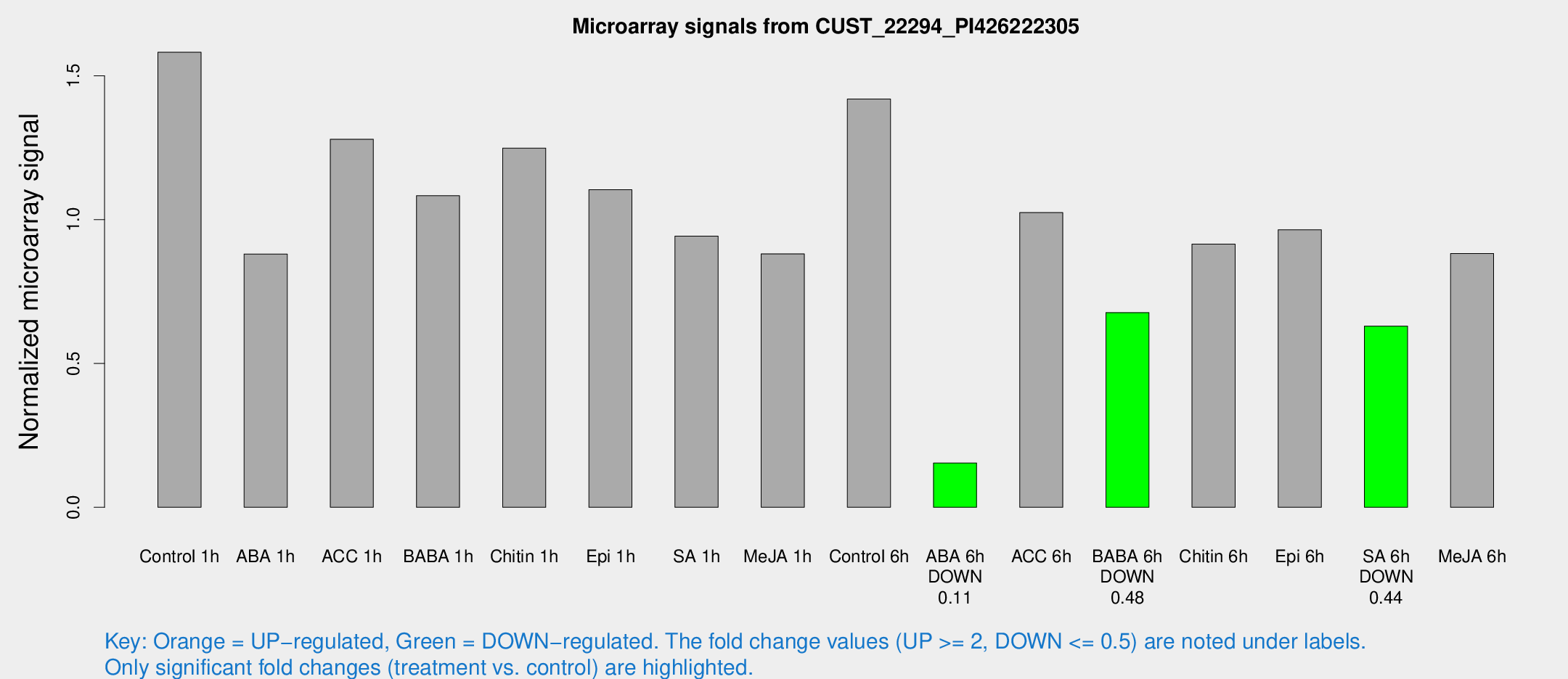
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 389.143 | 34.448 | 1.58203 | 0.092373 |
| ABA 1h | 191.808 | 17.8058 | 0.880604 | 0.0531515 |
| ACC 1h | 332.264 | 59.8482 | 1.27967 | 0.162425 |
| BABA 1h | 279.121 | 74.7757 | 1.08313 | 0.242306 |
| Chitin 1h | 278.917 | 36.9017 | 1.24869 | 0.141483 |
| Epi 1h | 234.631 | 19.0866 | 1.10405 | 0.107132 |
| SA 1h | 246.496 | 46.3291 | 0.942904 | 0.157941 |
| Me-JA 1h | 175.633 | 10.7465 | 0.881226 | 0.0561004 |
| Control 6h | 358.683 | 60.1888 | 1.41952 | 0.131209 |
| ABA 6h | 42.3476 | 9.14705 | 0.154051 | 0.0335025 |
| ACC 6h | 289.097 | 25.3633 | 1.0247 | 0.141545 |
| BABA 6h | 194.509 | 42.0107 | 0.676933 | 0.140381 |
| Chitin 6h | 242.225 | 31.9022 | 0.914744 | 0.135457 |
| Epi 6h | 283.372 | 74.4689 | 0.964523 | 0.285708 |
| SA 6h | 153.604 | 14.8971 | 0.630117 | 0.0398155 |
| Me-JA 6h | 231.992 | 64.9063 | 0.882331 | 0.179795 |
Source Transcript PGSC0003DMT400023394 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G22910.1 | +1 | 1e-76 | 253 | 127/190 (67%) | N-acetyl-l-glutamate synthase 1 | chr2:9749988-9752737 FORWARD LENGTH=609 |
