Probe CUST_22214_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22214_PI426222305 | JHI_St_60k_v1 | DMT400023398 | TTGGATATTCCAGCTCTGGAGAAGTTTTGAACTGCAAGAAGTGAGCCCAAGTATGTAGGC |
All Microarray Probes Designed to Gene DMG400009061
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22209_PI426222305 | JHI_St_60k_v1 | DMT400023396 | GGAAAGTGCAGCTGGTTTTTCCGACGAGAGCAGTGCAATAAAAGATGACTTATTCGTTGG |
| CUST_22294_PI426222305 | JHI_St_60k_v1 | DMT400023394 | TTTGTTGTTCTTCTGTCAGCTGAGATTATTGATAGCCCTTATTTGTCTTCCATAATCAAG |
| CUST_22314_PI426222305 | JHI_St_60k_v1 | DMT400023397 | TTCGTTGTTTTAATCTCAGCTGAAATTGTTGATAGTCCTCATTTGGATCATCTTCTCATG |
| CUST_22214_PI426222305 | JHI_St_60k_v1 | DMT400023398 | TTGGATATTCCAGCTCTGGAGAAGTTTTGAACTGCAAGAAGTGAGCCCAAGTATGTAGGC |
| CUST_22181_PI426222305 | JHI_St_60k_v1 | DMT400023399 | TTCGTTGTTTTAATCTCAGCTGAAATTGTTGATAGTCCTCATTTGGATCATCTTCTCATG |
Microarray Signals from CUST_22214_PI426222305
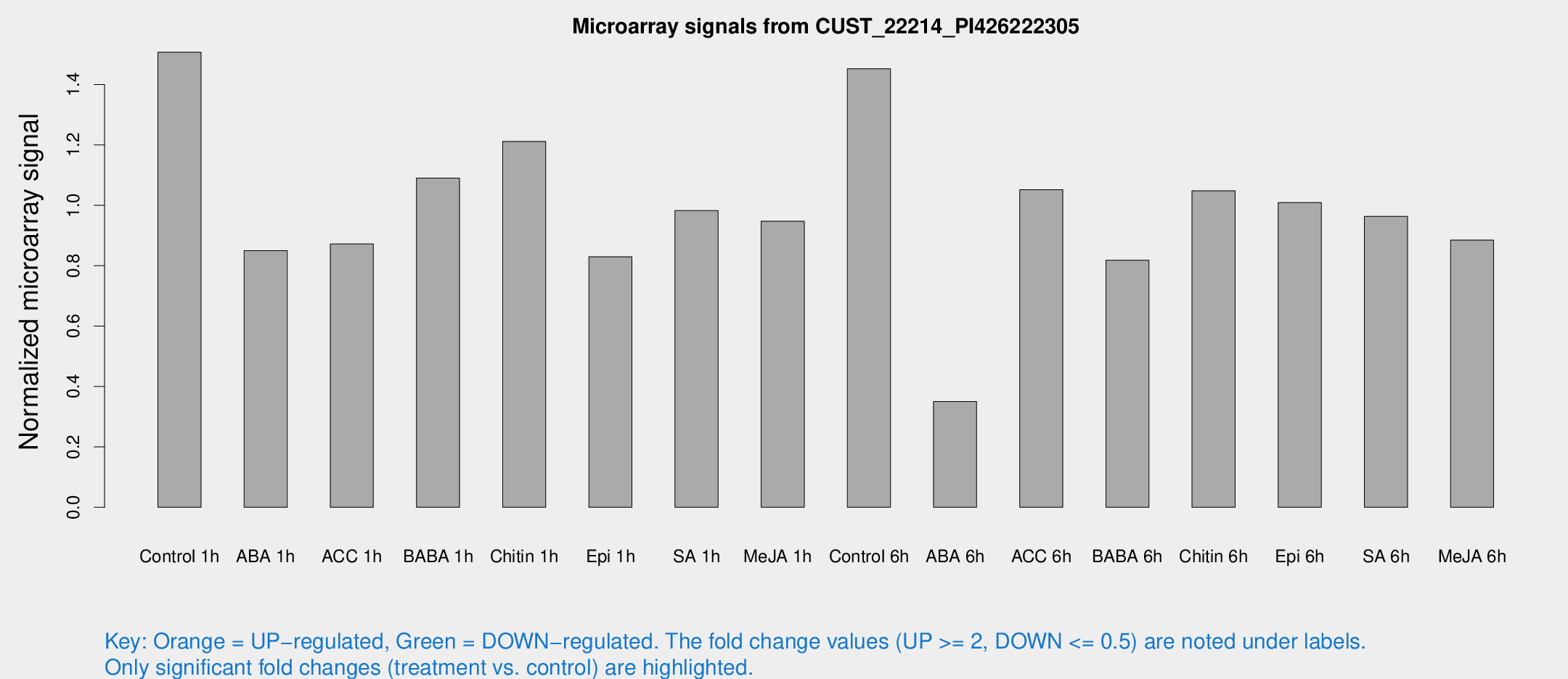
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 282.517 | 25.8466 | 1.50657 | 0.0892701 |
| ABA 1h | 140.129 | 8.84763 | 0.849549 | 0.0539884 |
| ACC 1h | 183.836 | 50.0747 | 0.872171 | 0.237606 |
| BABA 1h | 198.331 | 24.1989 | 1.08986 | 0.0667248 |
| Chitin 1h | 205.053 | 23.6441 | 1.21118 | 0.151943 |
| Epi 1h | 136.376 | 19.969 | 0.829278 | 0.122633 |
| SA 1h | 188.269 | 11.5175 | 0.982239 | 0.0628488 |
| Me-JA 1h | 144.919 | 12.435 | 0.947296 | 0.0602275 |
| Control 6h | 303.588 | 92.8868 | 1.45212 | 0.420218 |
| ABA 6h | 71.779 | 12.8415 | 0.35021 | 0.051604 |
| ACC 6h | 235.076 | 46.4509 | 1.05151 | 0.178145 |
| BABA 6h | 183.334 | 44.4747 | 0.818015 | 0.215909 |
| Chitin 6h | 217.087 | 42.4626 | 1.0478 | 0.247886 |
| Epi 6h | 226.425 | 60.7969 | 1.00907 | 0.300604 |
| SA 6h | 177.531 | 11.0854 | 0.963392 | 0.124744 |
| Me-JA 6h | 187.09 | 62.4032 | 0.884816 | 0.279644 |
Source Transcript PGSC0003DMT400023398 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G22910.1 | +2 | 0.0 | 703 | 358/488 (73%) | N-acetyl-l-glutamate synthase 1 | chr2:9749988-9752737 FORWARD LENGTH=609 |
