Probe CUST_22078_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22078_PI426222305 | JHI_St_60k_v1 | DMT400042003 | AATTGGAGGAGAAAGTATCACTAATGAAAGCAATAGCGCACTGTCACCGTCTCGGTGTAG |
All Microarray Probes Designed to Gene DMG400016291
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22105_PI426222305 | JHI_St_60k_v1 | DMT400042005 | GTGAAGCTAAGCGAAGTAGAATTCATAATCGGCTTTTATTTTAGTTTTTCGTCATGTCTG |
| CUST_22078_PI426222305 | JHI_St_60k_v1 | DMT400042003 | AATTGGAGGAGAAAGTATCACTAATGAAAGCAATAGCGCACTGTCACCGTCTCGGTGTAG |
| CUST_21978_PI426222305 | JHI_St_60k_v1 | DMT400042004 | TCAGAGTCCGAAGCTGTTGATGTCATGGTATCACTAATGAAAGCAATAGCGCACTGTCAC |
Microarray Signals from CUST_22078_PI426222305
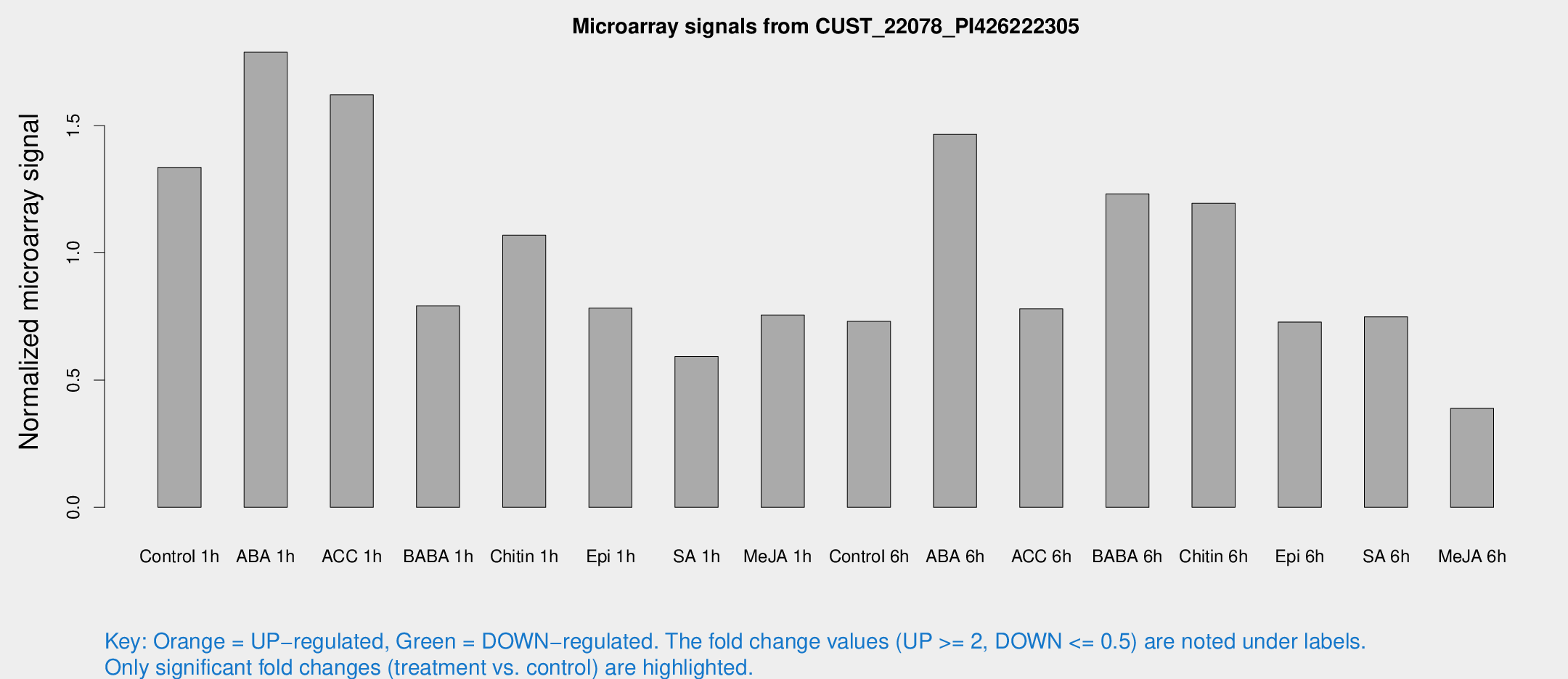
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 77.3926 | 17.1199 | 1.33569 | 0.200261 |
| ABA 1h | 88.9351 | 10.5138 | 1.78861 | 0.293473 |
| ACC 1h | 94.3975 | 14.1331 | 1.62121 | 0.276459 |
| BABA 1h | 42.8211 | 4.71673 | 0.791233 | 0.164129 |
| Chitin 1h | 63.2562 | 27.4906 | 1.06931 | 0.496957 |
| Epi 1h | 38.5446 | 5.90567 | 0.782604 | 0.124129 |
| SA 1h | 35.8868 | 8.3135 | 0.592867 | 0.203316 |
| Me-JA 1h | 34.5203 | 3.76094 | 0.755553 | 0.144334 |
| Control 6h | 73.3106 | 33.789 | 0.730838 | 1.25681 |
| ABA 6h | 87.7779 | 10.7481 | 1.46582 | 0.220436 |
| ACC 6h | 58.6777 | 25.2019 | 0.780035 | 0.217592 |
| BABA 6h | 80.3443 | 17.7988 | 1.23164 | 0.255401 |
| Chitin 6h | 71.5421 | 8.58577 | 1.19463 | 0.11318 |
| Epi 6h | 51.996 | 18.7384 | 0.728259 | 0.329984 |
| SA 6h | 43.2287 | 9.49962 | 0.748634 | 0.277095 |
| Me-JA 6h | 24.2431 | 7.65047 | 0.388824 | 0.129216 |
Source Transcript PGSC0003DMT400042003 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G08650.1 | +2 | 3e-89 | 275 | 151/324 (47%) | phosphoenolpyruvate carboxylase kinase 1 | chr1:2752206-2753232 FORWARD LENGTH=284 |
