Probe CUST_22017_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22017_PI426222305 | JHI_St_60k_v1 | DMT400042078 | TGACGCTTGCGTGAACTTTATTTGAAATGATCCAAGCCACCGATGTTTCTCATGTTCACG |
All Microarray Probes Designed to Gene DMG400016319
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22017_PI426222305 | JHI_St_60k_v1 | DMT400042078 | TGACGCTTGCGTGAACTTTATTTGAAATGATCCAAGCCACCGATGTTTCTCATGTTCACG |
| CUST_22032_PI426222305 | JHI_St_60k_v1 | DMT400042080 | TGACGCTTGCGTGAACTTTATTTGAAATGATCCAAGCCACCGATGTTTCTCATGTTCACG |
| CUST_22086_PI426222305 | JHI_St_60k_v1 | DMT400042079 | ATTTACTGCATACATTCACAGACTTCACTCTGTGTGATTACATTGGATATGTCTCGTTGT |
Microarray Signals from CUST_22017_PI426222305
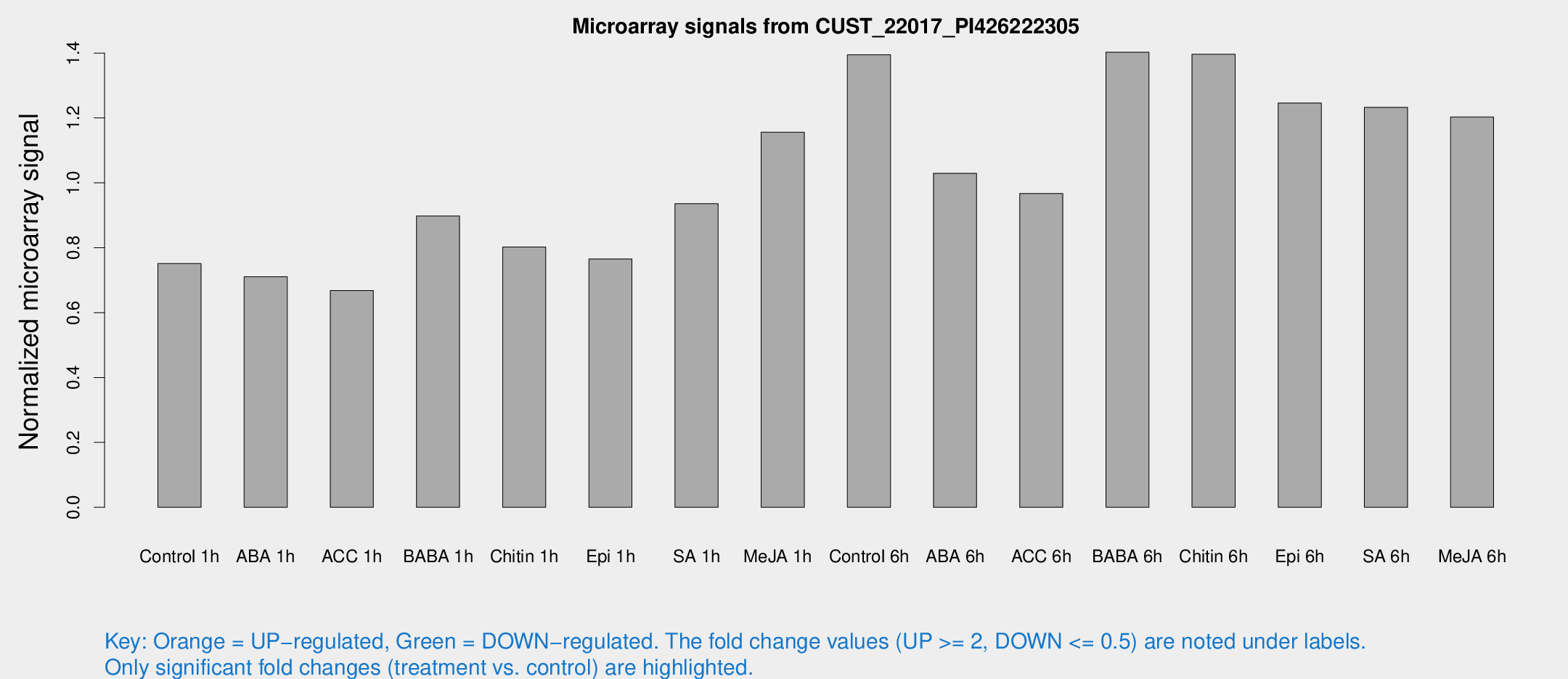
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3003.47 | 545.535 | 0.751315 | 0.11174 |
| ABA 1h | 2531.23 | 522.639 | 0.710444 | 0.0926629 |
| ACC 1h | 2807.1 | 631.579 | 0.66801 | 0.119847 |
| BABA 1h | 3512.24 | 706.489 | 0.89777 | 0.112498 |
| Chitin 1h | 2802.42 | 171.274 | 0.8022 | 0.0463242 |
| Epi 1h | 2616.71 | 352.204 | 0.765332 | 0.100141 |
| SA 1h | 3753.04 | 352.682 | 0.935663 | 0.0540266 |
| Me-JA 1h | 3693.02 | 391.722 | 1.15613 | 0.0761 |
| Control 6h | 5671.28 | 1119.4 | 1.39457 | 0.189141 |
| ABA 6h | 4275.94 | 423.748 | 1.02929 | 0.0594324 |
| ACC 6h | 4459.79 | 860.521 | 0.967163 | 0.078811 |
| BABA 6h | 6168.17 | 752.794 | 1.40254 | 0.143208 |
| Chitin 6h | 5792.59 | 482.036 | 1.39637 | 0.0806247 |
| Epi 6h | 5486.67 | 532.895 | 1.24585 | 0.14466 |
| SA 6h | 4741.5 | 322.153 | 1.23252 | 0.0834645 |
| Me-JA 6h | 4949.84 | 1169.96 | 1.20289 | 0.222685 |
Source Transcript PGSC0003DMT400042078 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G04280.1 | +1 | 8e-154 | 450 | 263/362 (73%) | P-loop containing nucleoside triphosphate hydrolases superfamily protein | chr1:1143643-1146541 REVERSE LENGTH=534 |
