Probe CUST_19793_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19793_PI426222305 | JHI_St_60k_v1 | DMT400064056 | CCTTTGACTTTGATGGTGCTTGGTAAATATACACACACATTCATTCTATCGCTAATTTGT |
All Microarray Probes Designed to Gene DMG400024895
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19793_PI426222305 | JHI_St_60k_v1 | DMT400064056 | CCTTTGACTTTGATGGTGCTTGGTAAATATACACACACATTCATTCTATCGCTAATTTGT |
| CUST_19881_PI426222305 | JHI_St_60k_v1 | DMT400064054 | GCTGGAGAAGTTGAGTCTCTTATGATGGAAAATTTAGCTTTGGATATGATAGACATTTCT |
| CUST_19721_PI426222305 | JHI_St_60k_v1 | DMT400064055 | CCACTCAAAAAGATGTCAACTCCAACTTTCAACCAAACAATGTCAGATTCCTTTTATGGA |
Microarray Signals from CUST_19793_PI426222305
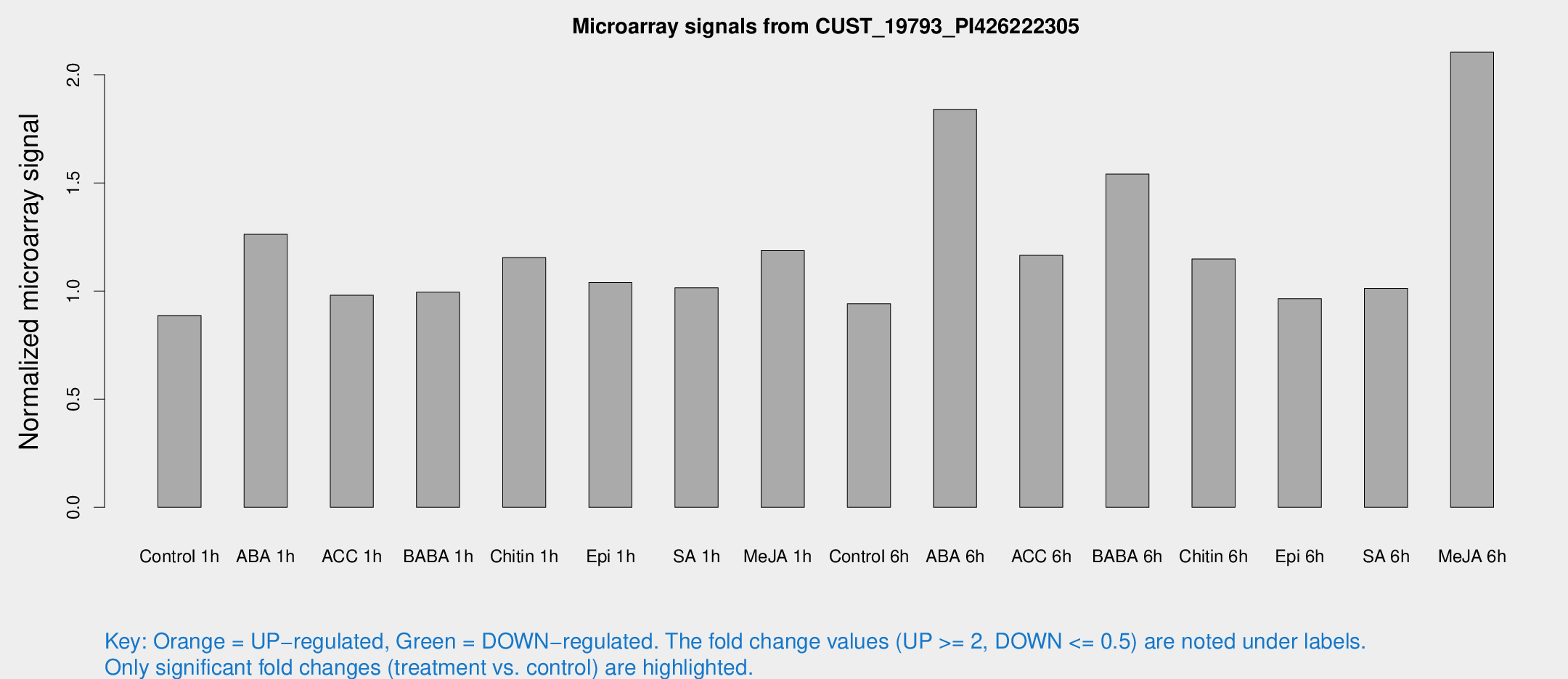
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.36803 | 3.6887 | 0.886816 | 0.51361 |
| ABA 1h | 8.37695 | 3.67978 | 1.26249 | 0.614859 |
| ACC 1h | 7.27384 | 4.23699 | 0.980996 | 0.568306 |
| BABA 1h | 6.88585 | 3.98821 | 0.994885 | 0.576169 |
| Chitin 1h | 7.6572 | 3.64628 | 1.15476 | 0.58037 |
| Epi 1h | 6.46212 | 3.68604 | 1.03923 | 0.592388 |
| SA 1h | 7.75101 | 3.728 | 1.01514 | 0.517582 |
| Me-JA 1h | 7.08398 | 3.79322 | 1.1871 | 0.645447 |
| Control 6h | 6.77597 | 3.93263 | 0.941588 | 0.546468 |
| ABA 6h | 16.019 | 5.29811 | 1.84017 | 0.928876 |
| ACC 6h | 9.98343 | 4.77254 | 1.16541 | 0.577211 |
| BABA 6h | 13.616 | 4.43811 | 1.54075 | 0.601395 |
| Chitin 6h | 9.01559 | 4.27723 | 1.14813 | 0.578275 |
| Epi 6h | 7.86289 | 4.59354 | 0.964732 | 0.559774 |
| SA 6h | 7.20129 | 4.07741 | 1.01235 | 0.574 |
| Me-JA 6h | 15.0885 | 3.90094 | 2.10432 | 0.545697 |
Source Transcript PGSC0003DMT400064056 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G80160.1 | +2 | 1e-40 | 122 | 56/92 (61%) | Lactoylglutathione lyase / glyoxalase I family protein | chr1:30151101-30151930 FORWARD LENGTH=167 |
