Probe CUST_19721_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19721_PI426222305 | JHI_St_60k_v1 | DMT400064055 | CCACTCAAAAAGATGTCAACTCCAACTTTCAACCAAACAATGTCAGATTCCTTTTATGGA |
All Microarray Probes Designed to Gene DMG400024895
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19793_PI426222305 | JHI_St_60k_v1 | DMT400064056 | CCTTTGACTTTGATGGTGCTTGGTAAATATACACACACATTCATTCTATCGCTAATTTGT |
| CUST_19881_PI426222305 | JHI_St_60k_v1 | DMT400064054 | GCTGGAGAAGTTGAGTCTCTTATGATGGAAAATTTAGCTTTGGATATGATAGACATTTCT |
| CUST_19721_PI426222305 | JHI_St_60k_v1 | DMT400064055 | CCACTCAAAAAGATGTCAACTCCAACTTTCAACCAAACAATGTCAGATTCCTTTTATGGA |
Microarray Signals from CUST_19721_PI426222305
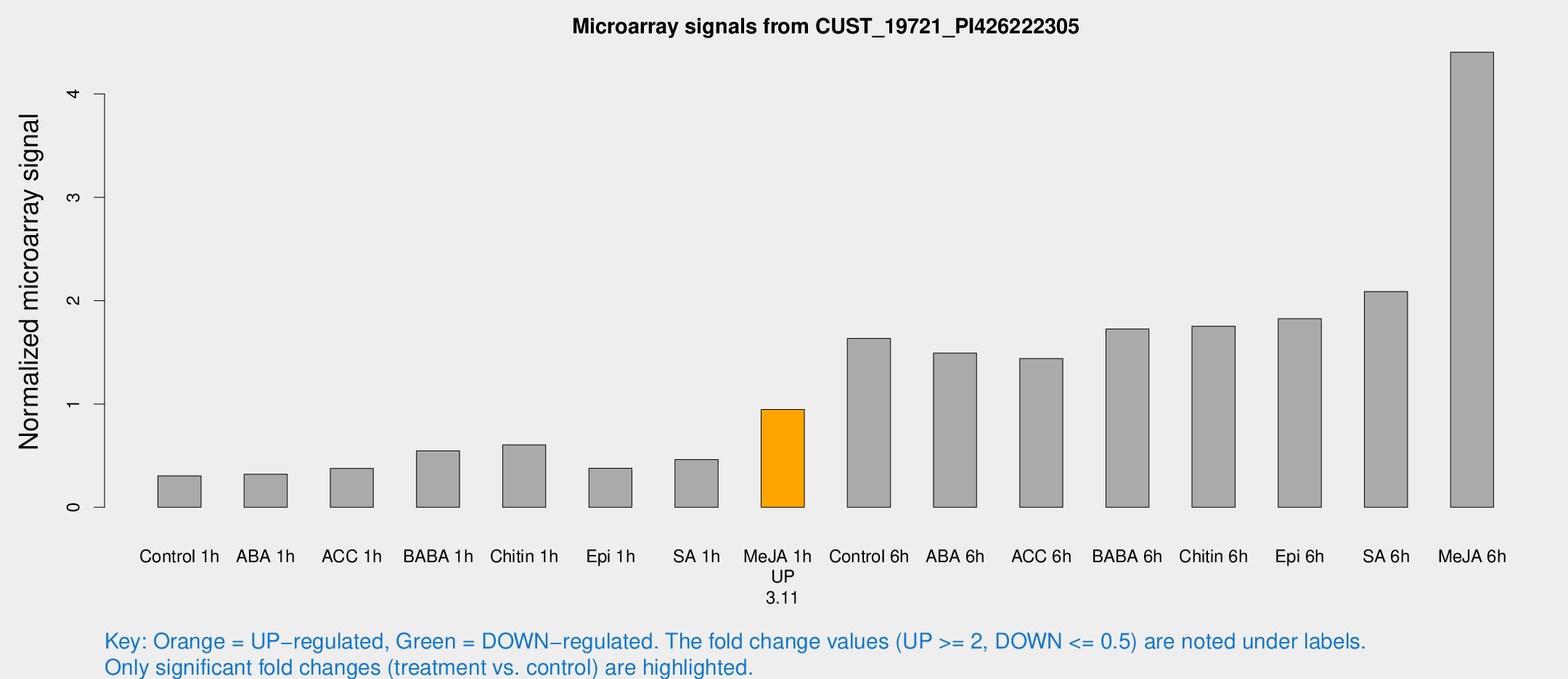
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 198.023 | 27.982 | 0.303907 | 0.0234647 |
| ABA 1h | 186.047 | 31.2808 | 0.32066 | 0.0302796 |
| ACC 1h | 250.66 | 29.3617 | 0.376552 | 0.0226175 |
| BABA 1h | 337.931 | 23.9882 | 0.546229 | 0.0917601 |
| Chitin 1h | 360.339 | 63.739 | 0.604882 | 0.0845488 |
| Epi 1h | 210.116 | 18.05 | 0.377322 | 0.0256697 |
| SA 1h | 329.263 | 97.4939 | 0.462628 | 0.14844 |
| Me-JA 1h | 493.921 | 28.7792 | 0.946309 | 0.0550662 |
| Control 6h | 1218.84 | 399.333 | 1.63548 | 0.580421 |
| ABA 6h | 1017.47 | 70.9454 | 1.49197 | 0.177607 |
| ACC 6h | 1072.86 | 137.109 | 1.44019 | 0.0923231 |
| BABA 6h | 1242.09 | 103.548 | 1.72528 | 0.0997711 |
| Chitin 6h | 1214.97 | 177.106 | 1.75256 | 0.248887 |
| Epi 6h | 1326.26 | 125.542 | 1.82667 | 0.19675 |
| SA 6h | 1333.09 | 133.537 | 2.08842 | 0.393131 |
| Me-JA 6h | 2879.25 | 479.306 | 4.40452 | 0.614146 |
Source Transcript PGSC0003DMT400064055 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G80160.1 | +3 | 3e-55 | 180 | 82/138 (59%) | Lactoylglutathione lyase / glyoxalase I family protein | chr1:30151101-30151930 FORWARD LENGTH=167 |
