Probe CUST_17947_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17947_PI426222305 | JHI_St_60k_v1 | DMT400018189 | GGAAGGGCTTTCTGGTTAGAATAGAAGATTTTTGTTCAGTTCTGAGTGTATTAAATAGTG |
All Microarray Probes Designed to Gene DMG400007058
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17947_PI426222305 | JHI_St_60k_v1 | DMT400018189 | GGAAGGGCTTTCTGGTTAGAATAGAAGATTTTTGTTCAGTTCTGAGTGTATTAAATAGTG |
| CUST_17928_PI426222305 | JHI_St_60k_v1 | DMT400018190 | GCCCTGTTGAGTTGGTATTTATGCCCCCTTGTATGTGTATATATATATATATATAGCCTA |
| CUST_18114_PI426222305 | JHI_St_60k_v1 | DMT400018187 | TATTCGTTGCCCAGATTGTTTAAAGTCTTCAAAAAGATGCAGCAAGATCAGCACAAGAAG |
| CUST_17986_PI426222305 | JHI_St_60k_v1 | DMT400018188 | TATTCGTTGCCCAGATTGTTTAAAGTCTTCAAAAAGATGCAGCAAGATCAGCACAAGAAG |
Microarray Signals from CUST_17947_PI426222305
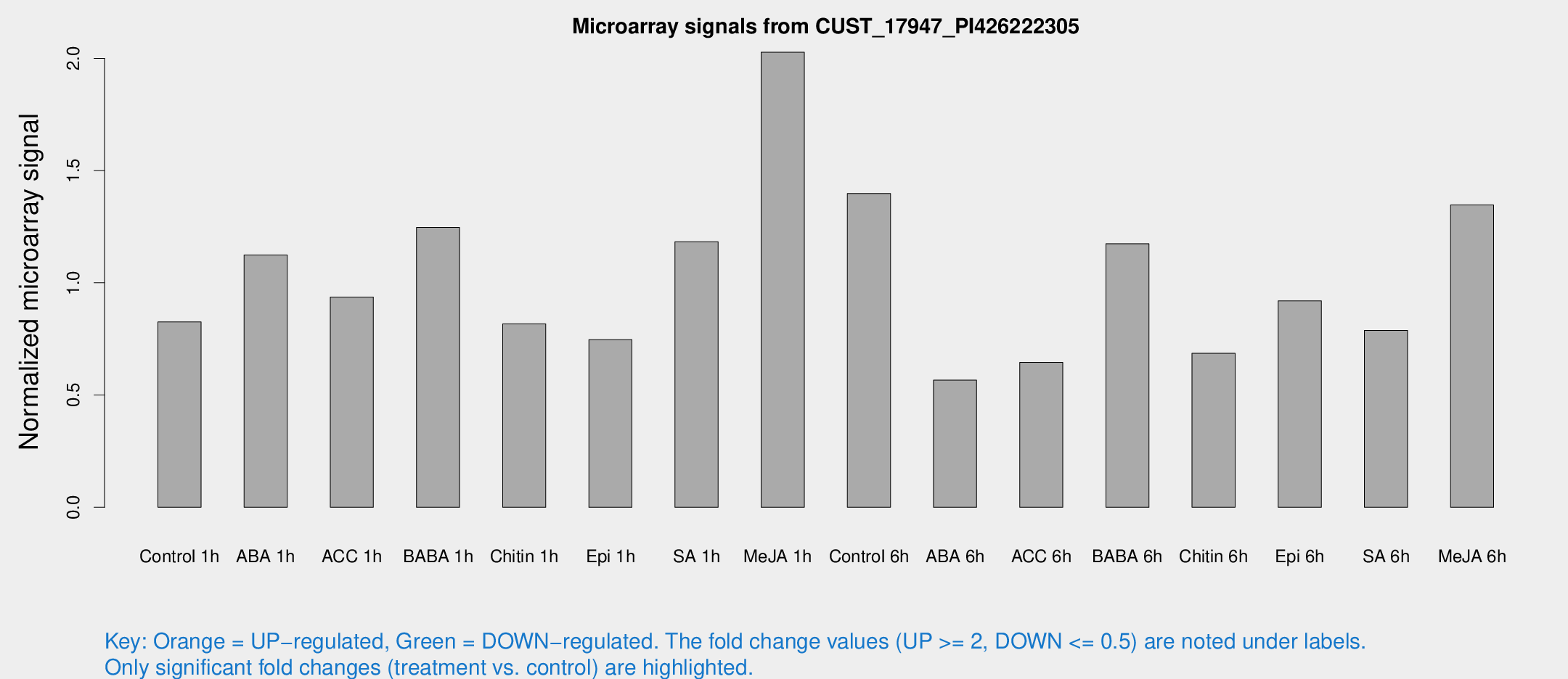
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 11.8629 | 4.88654 | 0.825541 | 0.386328 |
| ABA 1h | 13.6615 | 4.39109 | 1.12437 | 0.496115 |
| ACC 1h | 12.4374 | 4.82148 | 0.936786 | 0.400027 |
| BABA 1h | 15.6174 | 3.87828 | 1.24663 | 0.360388 |
| Chitin 1h | 9.75688 | 3.64627 | 0.817304 | 0.357877 |
| Epi 1h | 8.37698 | 3.55205 | 0.747129 | 0.353985 |
| SA 1h | 15.6888 | 3.71669 | 1.18264 | 0.317649 |
| Me-JA 1h | 21.7925 | 5.21499 | 2.02719 | 0.436973 |
| Control 6h | 20.1183 | 6.54784 | 1.39754 | 0.518867 |
| ABA 6h | 7.53571 | 3.76334 | 0.566323 | 0.293218 |
| ACC 6h | 9.30425 | 4.57719 | 0.645381 | 0.3216 |
| BABA 6h | 19.4371 | 6.76723 | 1.1743 | 0.60532 |
| Chitin 6h | 9.38546 | 4.14114 | 0.685877 | 0.335866 |
| Epi 6h | 13.1516 | 4.4377 | 0.919363 | 0.350745 |
| SA 6h | 9.99397 | 3.92828 | 0.787947 | 0.337602 |
| Me-JA 6h | 20.7477 | 8.21147 | 1.34662 | 0.744057 |
Source Transcript PGSC0003DMT400018189 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G14720.1 | +1 | 2e-138 | 422 | 257/408 (63%) | MAP kinase 19 | chr3:4946057-4948906 FORWARD LENGTH=598 |
