Probe CUST_17928_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17928_PI426222305 | JHI_St_60k_v1 | DMT400018190 | GCCCTGTTGAGTTGGTATTTATGCCCCCTTGTATGTGTATATATATATATATATAGCCTA |
All Microarray Probes Designed to Gene DMG400007058
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17947_PI426222305 | JHI_St_60k_v1 | DMT400018189 | GGAAGGGCTTTCTGGTTAGAATAGAAGATTTTTGTTCAGTTCTGAGTGTATTAAATAGTG |
| CUST_17928_PI426222305 | JHI_St_60k_v1 | DMT400018190 | GCCCTGTTGAGTTGGTATTTATGCCCCCTTGTATGTGTATATATATATATATATAGCCTA |
| CUST_18114_PI426222305 | JHI_St_60k_v1 | DMT400018187 | TATTCGTTGCCCAGATTGTTTAAAGTCTTCAAAAAGATGCAGCAAGATCAGCACAAGAAG |
| CUST_17986_PI426222305 | JHI_St_60k_v1 | DMT400018188 | TATTCGTTGCCCAGATTGTTTAAAGTCTTCAAAAAGATGCAGCAAGATCAGCACAAGAAG |
Microarray Signals from CUST_17928_PI426222305
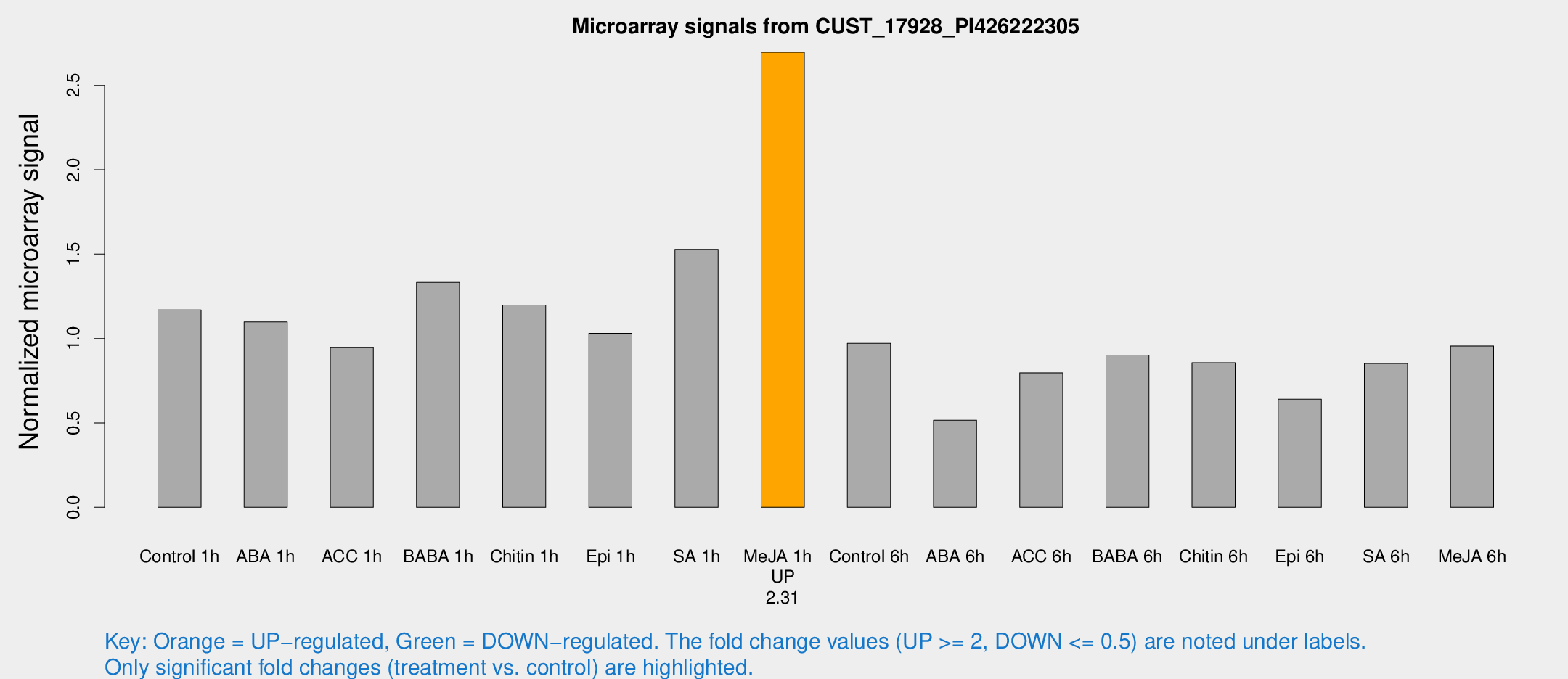
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6849.72 | 898.595 | 1.16971 | 0.111839 |
| ABA 1h | 5966.51 | 1548.37 | 1.09802 | 0.204006 |
| ACC 1h | 6036.53 | 1508.24 | 0.945555 | 0.211649 |
| BABA 1h | 7788.22 | 1656.68 | 1.33252 | 0.181903 |
| Chitin 1h | 6262.83 | 593.418 | 1.19846 | 0.111122 |
| Epi 1h | 5229.29 | 653.005 | 1.03111 | 0.120789 |
| SA 1h | 9065.91 | 524.902 | 1.52776 | 0.0882075 |
| Me-JA 1h | 12703.7 | 734.933 | 2.69626 | 0.170079 |
| Control 6h | 5807.69 | 1015.49 | 0.971413 | 0.104619 |
| ABA 6h | 3211.02 | 384.615 | 0.516367 | 0.0389152 |
| ACC 6h | 5371.89 | 763.652 | 0.797003 | 0.0460198 |
| BABA 6h | 5854.41 | 466.934 | 0.902013 | 0.0520822 |
| Chitin 6h | 5280.73 | 399.993 | 0.856494 | 0.0494549 |
| Epi 6h | 4196.2 | 381.136 | 0.64071 | 0.0456968 |
| SA 6h | 4910.96 | 542.896 | 0.852053 | 0.106894 |
| Me-JA 6h | 5664.3 | 948.698 | 0.956036 | 0.103425 |
Source Transcript PGSC0003DMT400018190 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G14720.1 | +1 | 7e-15 | 75 | 64/146 (44%) | MAP kinase 19 | chr3:4946057-4948906 FORWARD LENGTH=598 |
