Probe CUST_16847_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16847_PI426222305 | JHI_St_60k_v1 | DMT400068829 | ATGGATTCCGCTGATCCTGCTCGGGCATTTGTAAAGGATGTCAAACGAATCATTATTAAG |
All Microarray Probes Designed to Gene DMG402026767
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16828_PI426222305 | JHI_St_60k_v1 | DMT400068828 | AGATGCTTATGTGTACCTCACTCGTTGACAGTGCTCACACTGATAGCATCATTACTGAAG |
| CUST_16858_PI426222305 | JHI_St_60k_v1 | DMT400068830 | TTGATCATTGATATTTCTGAGACCATGAGTTACACGGGTTCTCTTTACTTGGATGTTGTT |
| CUST_16847_PI426222305 | JHI_St_60k_v1 | DMT400068829 | ATGGATTCCGCTGATCCTGCTCGGGCATTTGTAAAGGATGTCAAACGAATCATTATTAAG |
Microarray Signals from CUST_16847_PI426222305
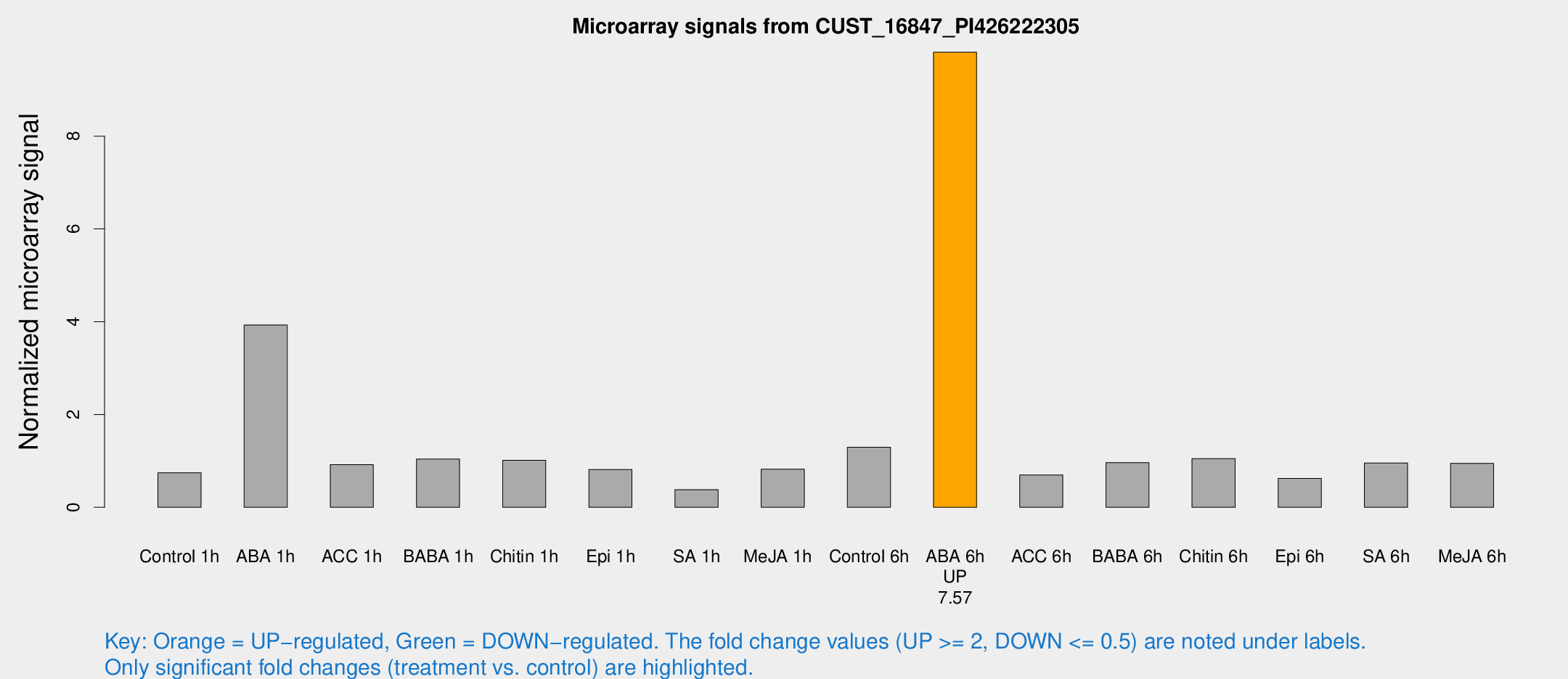
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 14.755 | 5.44158 | 0.7456 | 0.34008 |
| ABA 1h | 61.5331 | 14.5162 | 3.92993 | 0.927937 |
| ACC 1h | 16.1314 | 3.4351 | 0.921485 | 0.2087 |
| BABA 1h | 17.1154 | 3.27059 | 1.03988 | 0.21326 |
| Chitin 1h | 17.1194 | 5.98375 | 1.01389 | 0.362369 |
| Epi 1h | 12.5881 | 3.61431 | 0.815261 | 0.244322 |
| SA 1h | 6.61948 | 2.98123 | 0.380333 | 0.180112 |
| Me-JA 1h | 11.1843 | 3.10898 | 0.824025 | 0.229106 |
| Control 6h | 24.5048 | 7.53309 | 1.29629 | 0.403655 |
| ABA 6h | 178.48 | 29.9772 | 9.8093 | 1.46124 |
| ACC 6h | 13.4583 | 3.83529 | 0.696846 | 0.216043 |
| BABA 6h | 18.4845 | 3.61498 | 0.963664 | 0.202994 |
| Chitin 6h | 18.9193 | 3.5712 | 1.04953 | 0.2084 |
| Epi 6h | 12.7733 | 3.99304 | 0.622978 | 0.225774 |
| SA 6h | 19.6117 | 7.13462 | 0.955568 | 0.777481 |
| Me-JA 6h | 20.4213 | 8.29482 | 0.947679 | 0.573176 |
Source Transcript PGSC0003DMT400068829 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G39800.1 | +3 | 0.0 | 1012 | 550/717 (77%) | delta1-pyrroline-5-carboxylate synthase 1 | chr2:16598516-16602939 REVERSE LENGTH=717 |
