Probe CUST_16606_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16606_PI426222305 | JHI_St_60k_v1 | DMT400069484 | GTTCAATGTATTGATGAATTTGGTCTGTTCCTCTGTGAATTGAACCTTGTGTATTATTGC |
All Microarray Probes Designed to Gene DMG400027012
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16606_PI426222305 | JHI_St_60k_v1 | DMT400069484 | GTTCAATGTATTGATGAATTTGGTCTGTTCCTCTGTGAATTGAACCTTGTGTATTATTGC |
Microarray Signals from CUST_16606_PI426222305
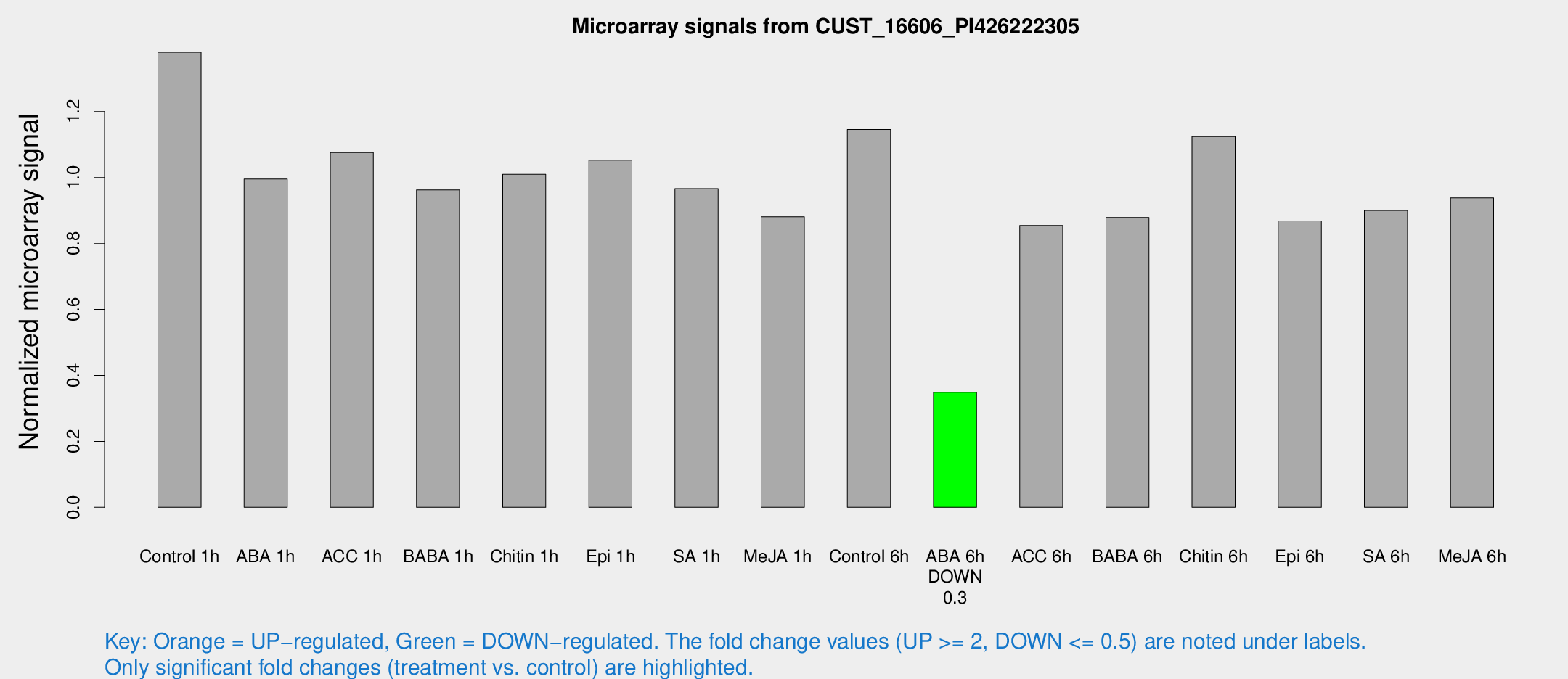
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 137068 | 13136.3 | 1.37994 | 0.079671 |
| ABA 1h | 87227.9 | 7231.21 | 0.995302 | 0.0765789 |
| ACC 1h | 110687 | 14220.6 | 1.07558 | 0.0672107 |
| BABA 1h | 97931.1 | 23195 | 0.962502 | 0.17139 |
| Chitin 1h | 90537.9 | 10005.8 | 1.00985 | 0.0583037 |
| Epi 1h | 89671.5 | 5179.81 | 1.05253 | 0.0607676 |
| SA 1h | 100462 | 16260.7 | 0.966133 | 0.10825 |
| Me-JA 1h | 71537.6 | 7078.52 | 0.881309 | 0.0508824 |
| Control 6h | 115555 | 17350.8 | 1.14548 | 0.112042 |
| ABA 6h | 36529.3 | 2111.51 | 0.348812 | 0.0303184 |
| ACC 6h | 97657 | 10570 | 0.85462 | 0.065855 |
| BABA 6h | 97049.4 | 5617.14 | 0.879065 | 0.0507528 |
| Chitin 6h | 118416 | 8847.75 | 1.12404 | 0.0648968 |
| Epi 6h | 98093.7 | 13500.3 | 0.86849 | 0.111903 |
| SA 6h | 92964.1 | 20301.7 | 0.900322 | 0.1247 |
| Me-JA 6h | 93430.2 | 11996.4 | 0.938177 | 0.0541657 |
Source Transcript PGSC0003DMT400069484 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G26850.1 | +3 | 0.0 | 630 | 312/406 (77%) | mannose-1-phosphate guanylyltransferase (GDP)s;GDP-galactose:mannose-1-phosphate guanylyltransferases;GDP-galactose:glucose-1-phosphate guanylyltransferases;GDP-galactose:myoinositol-1-phosphate guanylyltransferases;glucose-1-phosphate guanylyltransferase | chr4:13499262-13501145 REVERSE LENGTH=442 |
