Probe CUST_16504_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16504_PI426222305 | JHI_St_60k_v1 | DMT400069452 | GCACTGTACATAGAATATCCATTTTTTGTGTGATGATTGCTGAGTTGATACTAGGTGTTA |
All Microarray Probes Designed to Gene DMG400026998
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16504_PI426222305 | JHI_St_60k_v1 | DMT400069452 | GCACTGTACATAGAATATCCATTTTTTGTGTGATGATTGCTGAGTTGATACTAGGTGTTA |
Microarray Signals from CUST_16504_PI426222305
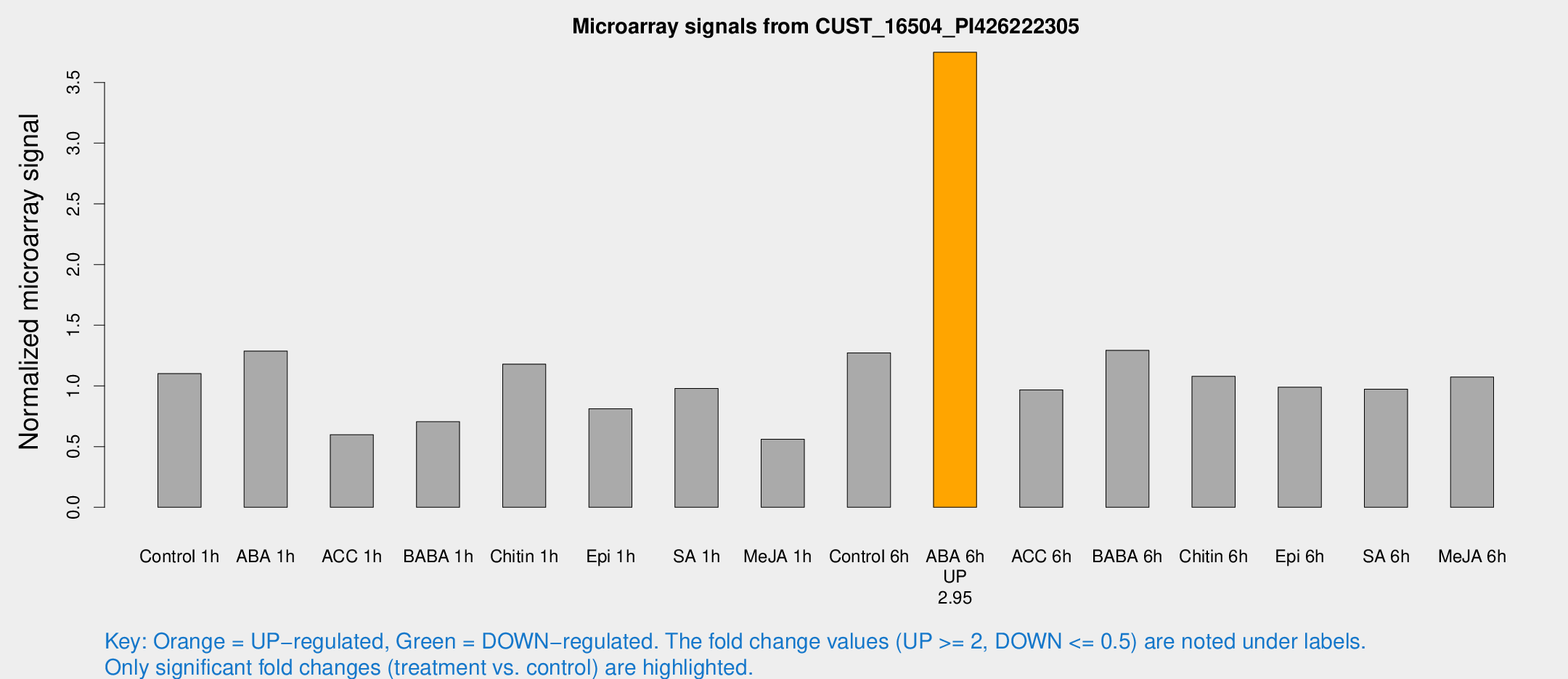
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 90.5309 | 11.2157 | 1.10083 | 0.120365 |
| ABA 1h | 92.3166 | 6.15303 | 1.2875 | 0.0857765 |
| ACC 1h | 50.416 | 5.59798 | 0.598626 | 0.0552831 |
| BABA 1h | 59.9555 | 15.2381 | 0.705611 | 0.148414 |
| Chitin 1h | 86.597 | 8.18343 | 1.17932 | 0.0809727 |
| Epi 1h | 58.1803 | 8.71675 | 0.81201 | 0.10123 |
| SA 1h | 81.7146 | 5.67219 | 0.979351 | 0.102024 |
| Me-JA 1h | 37.392 | 3.92325 | 0.560215 | 0.0592339 |
| Control 6h | 109.856 | 26.5478 | 1.27257 | 0.237913 |
| ABA 6h | 330.716 | 53.8165 | 3.7488 | 0.397354 |
| ACC 6h | 91.2028 | 11.3538 | 0.96765 | 0.0697148 |
| BABA 6h | 118.187 | 10.7857 | 1.29301 | 0.0853099 |
| Chitin 6h | 93.0072 | 6.52853 | 1.07863 | 0.0757728 |
| Epi 6h | 91.1313 | 8.14528 | 0.989856 | 0.0709748 |
| SA 6h | 86.8199 | 24.3914 | 0.972689 | 0.237802 |
| Me-JA 6h | 88.0105 | 11.1057 | 1.07319 | 0.0955594 |
Source Transcript PGSC0003DMT400069452 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G48020.1 | +3 | 9e-22 | 90 | 48/106 (45%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: chloroplast; EXPRESSED IN: 11 plant structures; EXPRESSED DURING: LP.04 four leaves visible, 4 anthesis; BEST Arabidopsis thaliana protein match is: unknown protein (TAIR:AT5G62865.1); Has 82 Blast hits to 82 proteins in 12 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 82; Viruses - 0; Other Eukaryotes - 0 (source: NCBI BLink). | chr3:17724593-17725000 FORWARD LENGTH=135 |
