Probe CUST_1463_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1463_PI426222305 | JHI_St_60k_v1 | DMT400032845 | GCTTCATCTGATCCTTTTTACGCAGTTATAGCTTATAGAAACTTTAAACCAGTGCATGAA |
All Microarray Probes Designed to Gene DMG400012615
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1463_PI426222305 | JHI_St_60k_v1 | DMT400032845 | GCTTCATCTGATCCTTTTTACGCAGTTATAGCTTATAGAAACTTTAAACCAGTGCATGAA |
Microarray Signals from CUST_1463_PI426222305
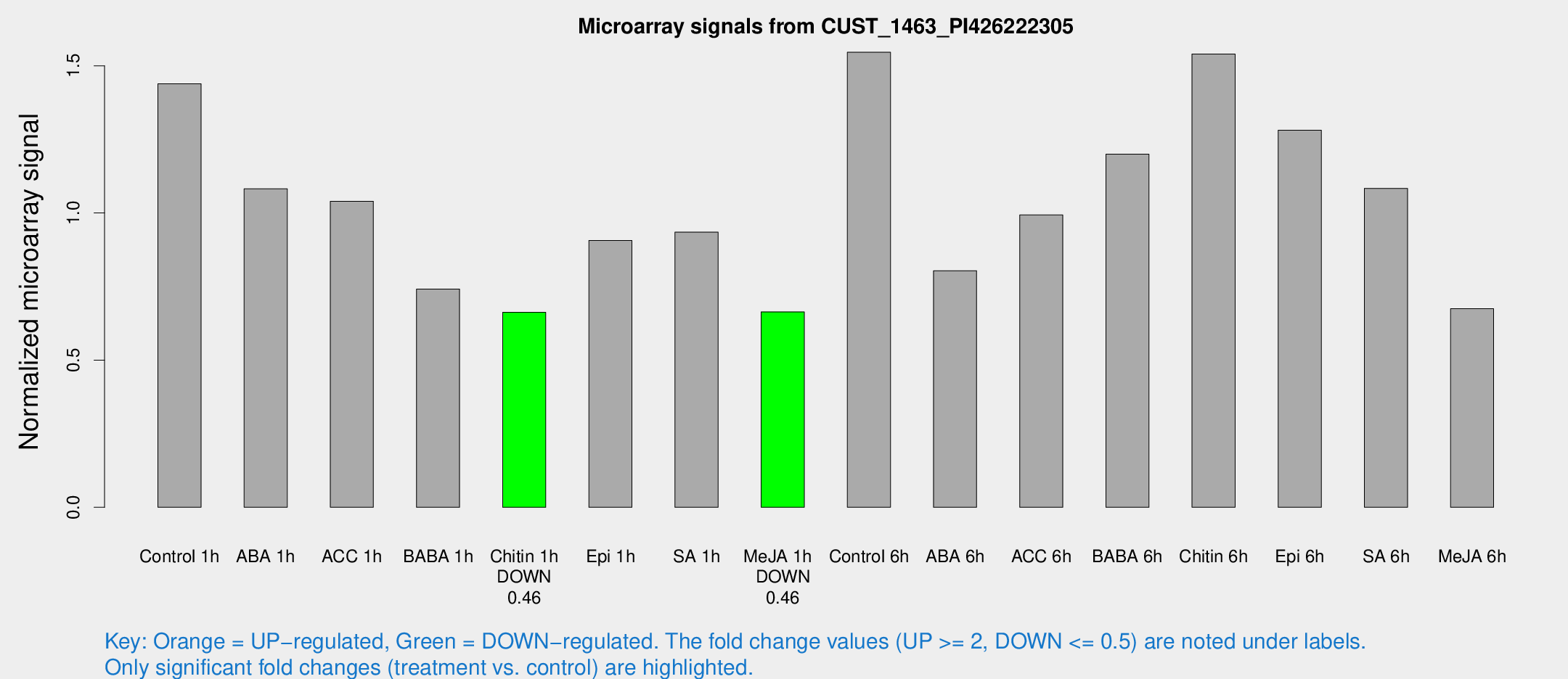
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3583.77 | 435.557 | 1.43879 | 0.0830793 |
| ABA 1h | 2357.84 | 136.552 | 1.08212 | 0.0624928 |
| ACC 1h | 2774.54 | 591.227 | 1.03968 | 0.180465 |
| BABA 1h | 1838.1 | 358.27 | 0.741528 | 0.0877236 |
| Chitin 1h | 1463.51 | 84.5953 | 0.66234 | 0.0421249 |
| Epi 1h | 1934.62 | 117.171 | 0.906593 | 0.0527303 |
| SA 1h | 2370.34 | 166.392 | 0.934665 | 0.0539774 |
| Me-JA 1h | 1334.94 | 77.366 | 0.663845 | 0.0383619 |
| Control 6h | 4111.04 | 1032.82 | 1.54617 | 0.326214 |
| ABA 6h | 2104.82 | 121.911 | 0.803773 | 0.0464276 |
| ACC 6h | 2956.13 | 685.128 | 0.99328 | 0.116336 |
| BABA 6h | 3440.03 | 718.365 | 1.19968 | 0.263984 |
| Chitin 6h | 4062.41 | 382.663 | 1.54014 | 0.120724 |
| Epi 6h | 3572.77 | 295.074 | 1.28143 | 0.162361 |
| SA 6h | 2782.09 | 594.497 | 1.08369 | 0.141352 |
| Me-JA 6h | 1815.55 | 527.296 | 0.67471 | 0.163795 |
Source Transcript PGSC0003DMT400032845 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc02g093550.2 | +1 | 0.0 | 538 | 264/291 (91%) | genomic_reference:SL2.50ch02 gene_region:48960510-48961856 transcript_region:SL2.50ch02:48960510..48961856+ go_terms:GO:0008152 functional_description:Methyltransferase type 11 (AHRD V1 ***- C7QSM0_CYAP0); contains Interpro domain(s) IPR013216 Methyltransferase type 11 |
| TAIR PP10 | AT3G01660.1 | +1 | 8e-116 | 340 | 173/279 (62%) | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | chr3:245532-246432 FORWARD LENGTH=273 |
