Probe CUST_13737_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13737_PI426222305 | JHI_St_60k_v1 | DMT400084204 | TTGGGCTGCCATGCAGTTGGCATAATAATCTTGGTTAGCGTACCTCCAACAACAACTACA |
All Microarray Probes Designed to Gene DMG400033876
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13737_PI426222305 | JHI_St_60k_v1 | DMT400084204 | TTGGGCTGCCATGCAGTTGGCATAATAATCTTGGTTAGCGTACCTCCAACAACAACTACA |
| CUST_13652_PI426222305 | JHI_St_60k_v1 | DMT400084203 | GATTTGTTTTATGCAGTTGGCATAATAATCTTGGTTAGCGTACCTCCAACAACAACTACA |
| CUST_13740_PI426222305 | JHI_St_60k_v1 | DMT400084206 | TACCGGGTGTATTGACAGGATCAAAGCAATTTTCATGTGTATCCTGATTGGGCTGCCATG |
Microarray Signals from CUST_13737_PI426222305
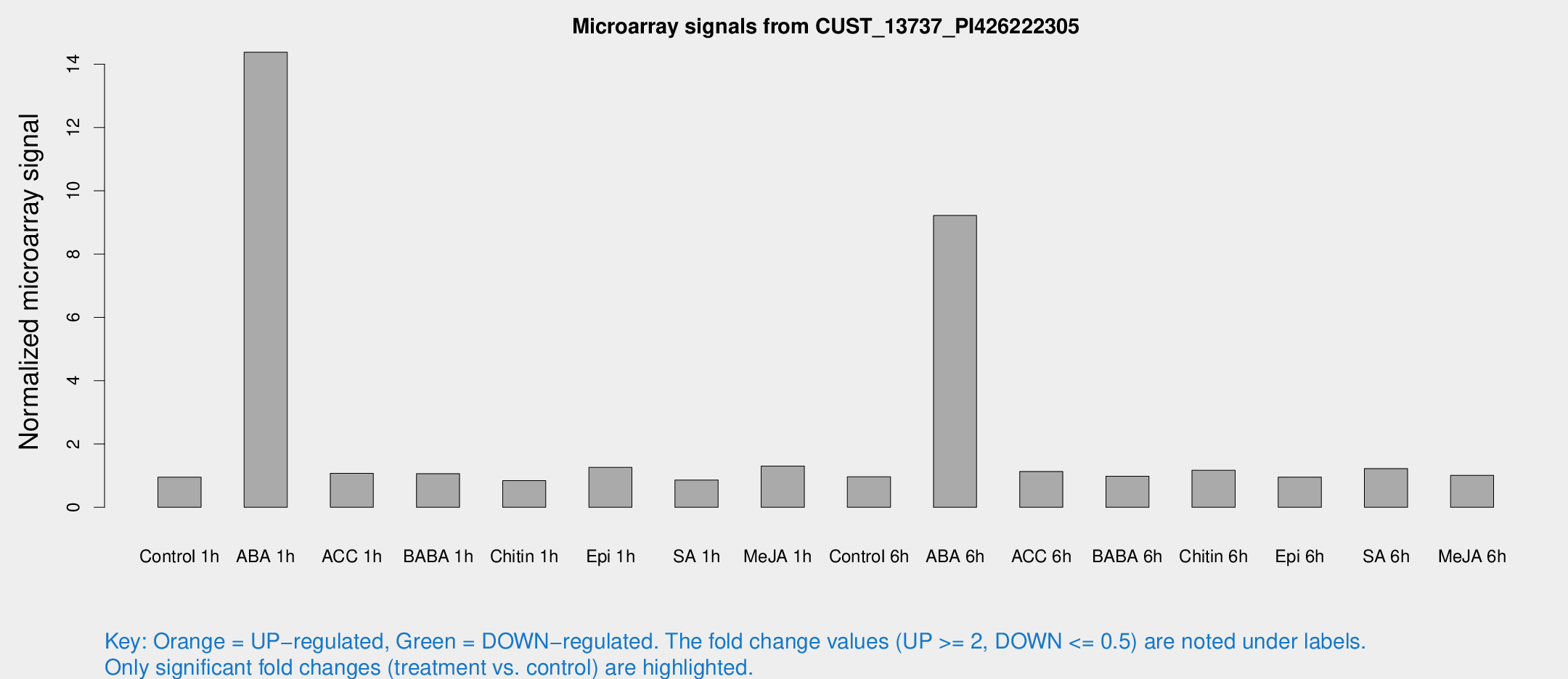
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7.3884 | 3.09529 | 0.952348 | 0.461894 |
| ABA 1h | 98.7251 | 29.1597 | 14.3758 | 4.32975 |
| ACC 1h | 8.30117 | 3.40728 | 1.0748 | 0.491914 |
| BABA 1h | 8.14189 | 3.26191 | 1.06345 | 0.509931 |
| Chitin 1h | 5.32664 | 3.06154 | 0.843368 | 0.470543 |
| Epi 1h | 8.14692 | 3.07153 | 1.26223 | 0.514128 |
| SA 1h | 6.61157 | 3.05763 | 0.860263 | 0.430772 |
| Me-JA 1h | 7.92175 | 3.13179 | 1.3024 | 0.552303 |
| Control 6h | 7.31631 | 3.15646 | 0.963071 | 0.467897 |
| ABA 6h | 80.4467 | 25.6919 | 9.21803 | 3.62035 |
| ACC 6h | 9.39732 | 3.81845 | 1.13192 | 0.469779 |
| BABA 6h | 8.18723 | 3.49447 | 0.983577 | 0.445769 |
| Chitin 6h | 10.6605 | 4.70552 | 1.16843 | 0.584473 |
| Epi 6h | 8.14304 | 3.62547 | 0.953248 | 0.466445 |
| SA 6h | 9.41713 | 3.32407 | 1.2245 | 0.511333 |
| Me-JA 6h | 8.05574 | 3.11466 | 1.01223 | 0.480552 |
Source Transcript PGSC0003DMT400084204 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G46270.1 | +2 | 7e-68 | 231 | 187/442 (42%) | G-box binding factor 3 | chr2:19000859-19002901 FORWARD LENGTH=382 |
