Probe CUST_13652_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13652_PI426222305 | JHI_St_60k_v1 | DMT400084203 | GATTTGTTTTATGCAGTTGGCATAATAATCTTGGTTAGCGTACCTCCAACAACAACTACA |
All Microarray Probes Designed to Gene DMG400033876
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13737_PI426222305 | JHI_St_60k_v1 | DMT400084204 | TTGGGCTGCCATGCAGTTGGCATAATAATCTTGGTTAGCGTACCTCCAACAACAACTACA |
| CUST_13652_PI426222305 | JHI_St_60k_v1 | DMT400084203 | GATTTGTTTTATGCAGTTGGCATAATAATCTTGGTTAGCGTACCTCCAACAACAACTACA |
| CUST_13740_PI426222305 | JHI_St_60k_v1 | DMT400084206 | TACCGGGTGTATTGACAGGATCAAAGCAATTTTCATGTGTATCCTGATTGGGCTGCCATG |
Microarray Signals from CUST_13652_PI426222305
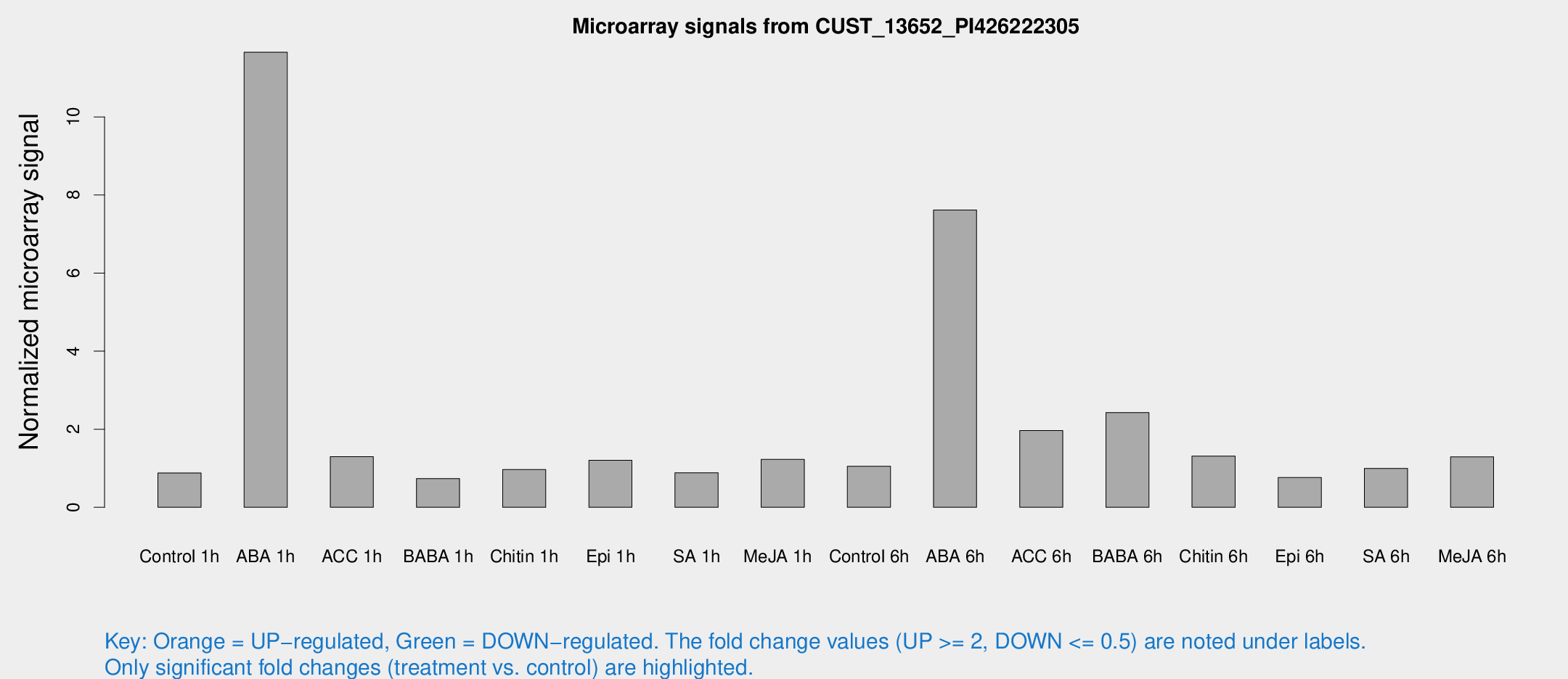
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.99217 | 3.1196 | 0.878101 | 0.402604 |
| ABA 1h | 91.0015 | 32.6862 | 11.6562 | 4.34609 |
| ACC 1h | 10.5959 | 3.46332 | 1.29813 | 0.438457 |
| BABA 1h | 5.59319 | 3.24731 | 0.737049 | 0.427074 |
| Chitin 1h | 7.10764 | 3.14731 | 0.967891 | 0.470897 |
| Epi 1h | 8.66796 | 3.0995 | 1.20342 | 0.49784 |
| SA 1h | 8.079 | 3.09919 | 0.885293 | 0.421376 |
| Me-JA 1h | 8.91 | 3.25783 | 1.22908 | 0.574616 |
| Control 6h | 8.57568 | 3.31764 | 1.05246 | 0.444562 |
| ABA 6h | 82.9044 | 33.558 | 7.61605 | 5.17345 |
| ACC 6h | 43.1752 | 36.0717 | 1.96622 | 4.75069 |
| BABA 6h | 28.8418 | 13.3678 | 2.42691 | 2.06097 |
| Chitin 6h | 11.7038 | 3.646 | 1.31211 | 0.489359 |
| Epi 6h | 6.8352 | 3.7476 | 0.763148 | 0.416513 |
| SA 6h | 8.46962 | 3.43144 | 0.99778 | 0.478642 |
| Me-JA 6h | 11.073 | 3.31859 | 1.29219 | 0.477785 |
Source Transcript PGSC0003DMT400084203 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G46270.1 | +1 | 3e-73 | 245 | 187/415 (45%) | G-box binding factor 3 | chr2:19000859-19002901 FORWARD LENGTH=382 |
