Probe CUST_12318_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12318_PI426222305 | JHI_St_60k_v1 | DMT400063777 | GCCCCATTGTACATTTTCTGTCATTACACTTCGTGGTATATAAACATATAGCTGTACATT |
All Microarray Probes Designed to Gene DMG400024785
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12612_PI426222305 | JHI_St_60k_v1 | DMT400063775 | GCCCCATTGTACATTTTCTGTCATTACACTTCGTGGTATATAAACATATAGCTGTACATT |
| CUST_12318_PI426222305 | JHI_St_60k_v1 | DMT400063777 | GCCCCATTGTACATTTTCTGTCATTACACTTCGTGGTATATAAACATATAGCTGTACATT |
| CUST_12536_PI426222305 | JHI_St_60k_v1 | DMT400063776 | GCCCCATTGTACATTTTCTGTCATTACACTTCGTGGTATATAAACATATAGCTGTACATT |
Microarray Signals from CUST_12318_PI426222305
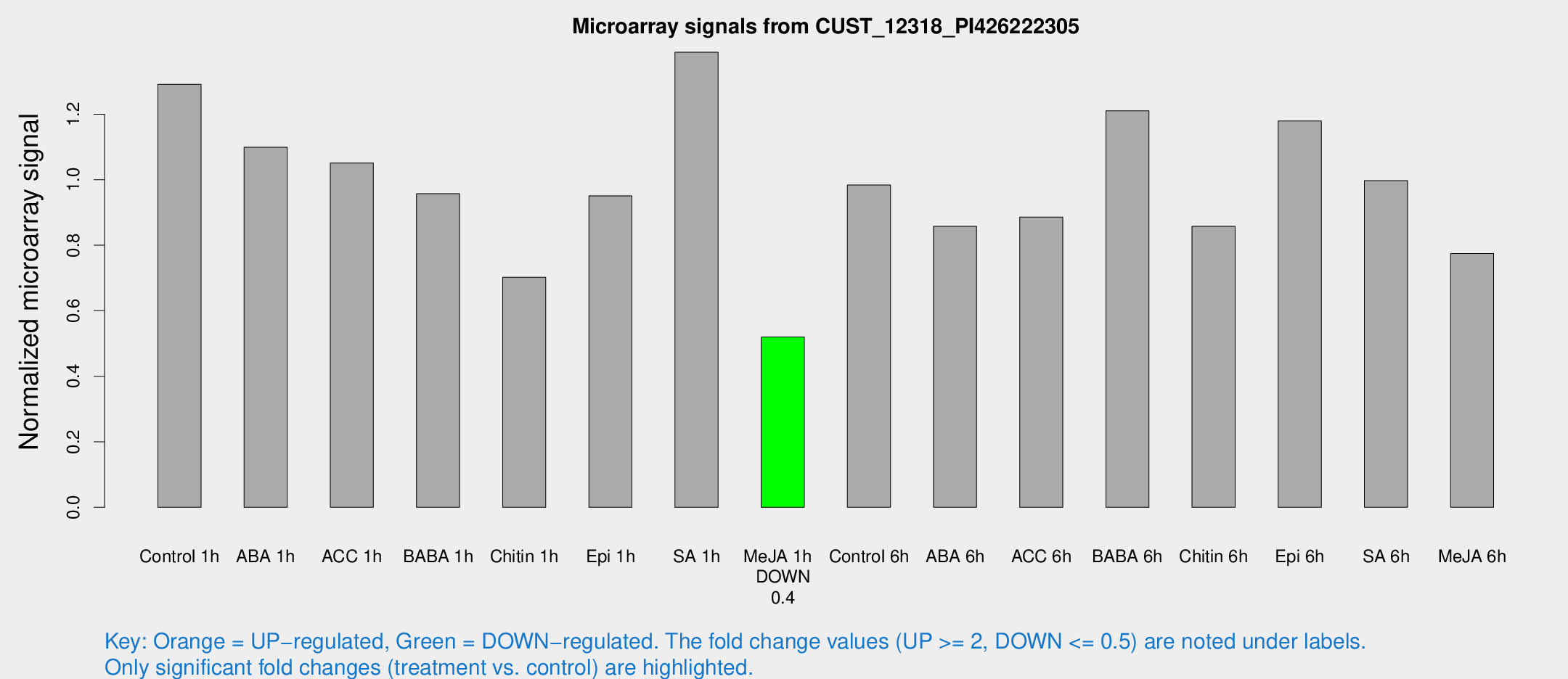
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 380.89 | 83.0616 | 1.29126 | 0.196419 |
| ABA 1h | 276.716 | 24.43 | 1.09929 | 0.0649838 |
| ACC 1h | 357.793 | 115.717 | 1.05094 | 0.419572 |
| BABA 1h | 267.676 | 41.3781 | 0.957293 | 0.0635765 |
| Chitin 1h | 179.682 | 15.5448 | 0.702289 | 0.0429374 |
| Epi 1h | 232.833 | 13.9139 | 0.95074 | 0.0567465 |
| SA 1h | 405.848 | 33.046 | 1.38911 | 0.0846956 |
| Me-JA 1h | 121.834 | 15.5008 | 0.51982 | 0.05818 |
| Control 6h | 289.628 | 52.8099 | 0.98377 | 0.108246 |
| ABA 6h | 272.317 | 61.9206 | 0.857497 | 0.16409 |
| ACC 6h | 288.479 | 20.708 | 0.885869 | 0.0600901 |
| BABA 6h | 383.036 | 22.5038 | 1.21015 | 0.0710248 |
| Chitin 6h | 258.691 | 15.5091 | 0.857907 | 0.0513433 |
| Epi 6h | 377.992 | 30.4697 | 1.17906 | 0.0792596 |
| SA 6h | 291.515 | 57.2532 | 0.997178 | 0.106642 |
| Me-JA 6h | 236.255 | 67.9284 | 0.774519 | 0.160304 |
Source Transcript PGSC0003DMT400063777 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G18750.1 | +1 | 1e-79 | 273 | 158/361 (44%) | Calmodulin-binding protein | chr2:8125827-8128363 FORWARD LENGTH=622 |
