Probe CUST_1082_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1082_PI426222305 | JHI_St_60k_v1 | DMT400089418 | CGGAGGGAGCTCAGAAAAGAGGCTGAGGAAATTCATCGAAACATTAACAGCAGAACAAAA |
All Microarray Probes Designed to Gene DMG400038989
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1082_PI426222305 | JHI_St_60k_v1 | DMT400089418 | CGGAGGGAGCTCAGAAAAGAGGCTGAGGAAATTCATCGAAACATTAACAGCAGAACAAAA |
Microarray Signals from CUST_1082_PI426222305
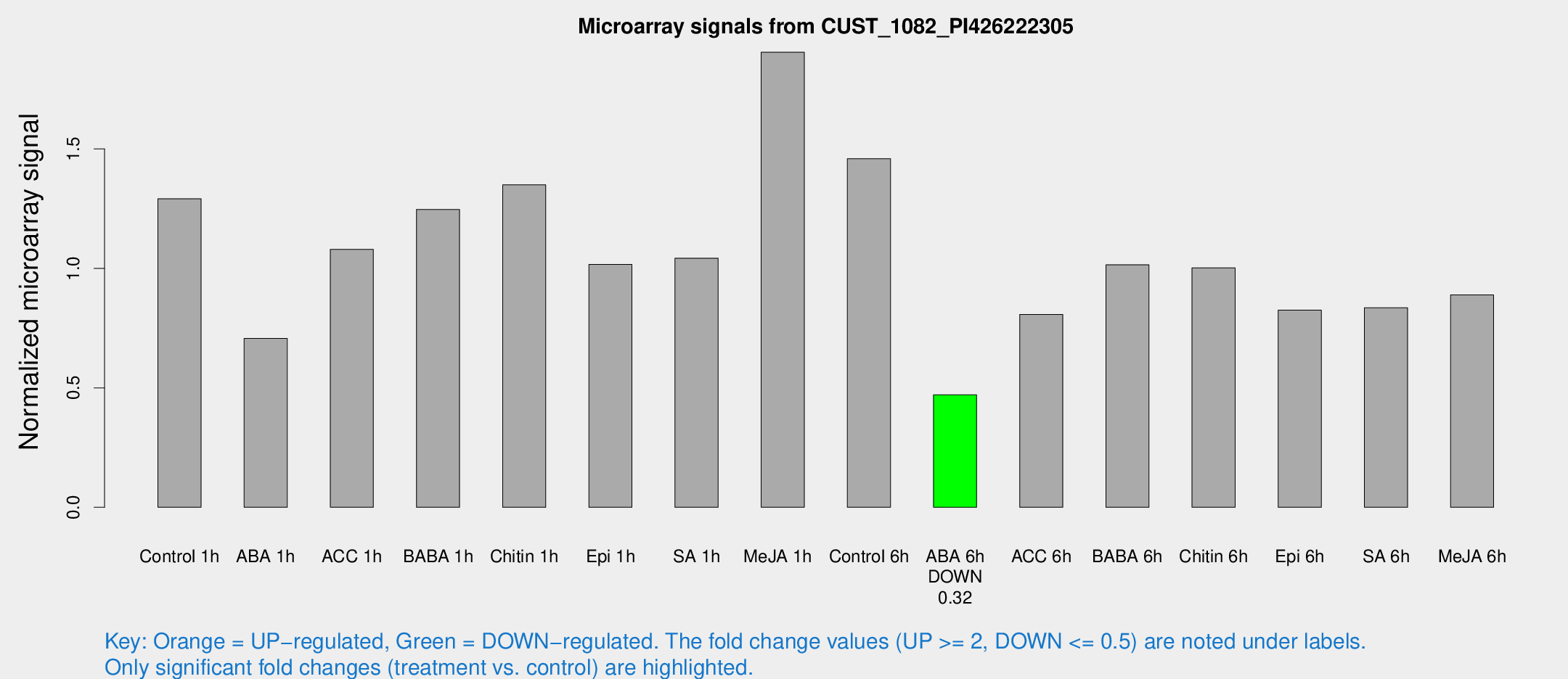
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 99.2198 | 6.42534 | 1.29124 | 0.0835835 |
| ABA 1h | 51.5104 | 13.4742 | 0.706777 | 0.233555 |
| ACC 1h | 85.7475 | 7.80564 | 1.07936 | 0.07511 |
| BABA 1h | 93.0359 | 8.89751 | 1.24647 | 0.249578 |
| Chitin 1h | 93.2745 | 6.12062 | 1.3494 | 0.0882962 |
| Epi 1h | 68.4874 | 7.97286 | 1.01621 | 0.109962 |
| SA 1h | 82.2174 | 5.56355 | 1.04264 | 0.0789406 |
| Me-JA 1h | 119.406 | 7.50088 | 1.90455 | 0.225719 |
| Control 6h | 124.42 | 35.8631 | 1.4588 | 0.389035 |
| ABA 6h | 38.457 | 3.80548 | 0.470849 | 0.0465529 |
| ACC 6h | 74.6847 | 15.5482 | 0.807265 | 0.119423 |
| BABA 6h | 88.3762 | 10.6908 | 1.01471 | 0.0840404 |
| Chitin 6h | 81.8857 | 5.75766 | 1.00214 | 0.0704441 |
| Epi 6h | 71.5363 | 5.3758 | 0.825059 | 0.0617068 |
| SA 6h | 69.3672 | 19.4907 | 0.834886 | 0.39517 |
| Me-JA 6h | 69.1091 | 9.39658 | 0.888997 | 0.121057 |
Source Transcript PGSC0003DMT400089418 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc02g088500.1 | +1 | 0.0 | 839 | 444/463 (96%) | evidence_code:10F0H1E1IEG genomic_reference:SL2.50ch02 gene_region:45151406-45152794 transcript_region:SL2.50ch02:45151406..45152794- go_terms:GO:0080043 functional_description:C-glucosyltransferase (AHRD V1 **** C3W7B0_ORYSJ); contains Interpro domain(s) IPR002213 UDP-glucuronosyl/UDP-glucosyltransferase |
| TAIR PP10 | AT4G36770.1 | +1 | 2e-35 | 137 | 123/448 (27%) | UDP-Glycosyltransferase superfamily protein | chr4:17330217-17331590 REVERSE LENGTH=457 |
